miR-22 Regulates Invasion, Gene Expression and Predicts Overall Survival in Patients with Clear Cell Renal Cell Carcinoma
- PMID: 31763513
- PMCID: PMC6839454
- DOI: 10.3233/KCA-190051
miR-22 Regulates Invasion, Gene Expression and Predicts Overall Survival in Patients with Clear Cell Renal Cell Carcinoma
Abstract
Background: Clear cell renal cell carcinoma (ccRCC) is molecularly diverse and distinct molecular subtypes show different clinical outcomes. MicroRNAs (miRNAs) are essential components of gene regulatory networks and play a crucial role in progression of many cancer types including ccRCC.
Objective: Identify prognostic miRNAs and determine the role of miR-22 in ccRCC.
Methods: Hierarchical clustering was done in R using gene expression profiles of over 450 ccRCC cases in The Cancer Genome Atlas (TCGA). Kaplan-Meier analysis was performed to identify prognostic miRNAs in the TCGA dataset. RNA-Seq was performed to identify miR-22 target genes in primary ccRCC cells and Matrigel invasion assay was performed to assess the effects of miR-22 overexpression on cell invasion.
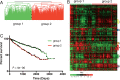
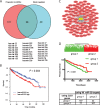
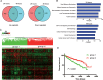
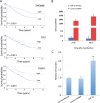
Results: Hierarchical clustering analysis using 2,621 prognostic genes previously identified by our group demonstrated that ccRCC patients with longer overall survival expressed lower levels of genes promoting proliferation or immune responses, while better maintaining gene expression associated with cortical differentiation and cell adhesion. Targets of 26 miRNAs were significantly enriched in the 2,621 prognostic genes and these miRNAs were prognostic by themselves. MiR-22 was associated with poor overall survival in the TCGA dataset. Overexpression of miR-22 promoted invasion of primary ccRCC cells in vitro and modulated transcriptional programs implicated in cancer progression including DNA repair, cell proliferation and invasion.
Conclusions: Our results suggest that ccRCCs with differential clinical outcomes have distinct transcriptomes for which miRNAs could serve as master regulators. MiR-22, as a master regulator, promotes ccRCC progression at least in part by enhancing cell invasion.
Keywords: MicroRNA; TCGA; clear cell renal cell carcinoma; miR-22; survival.
© 2019 – IOS Press and the authors. All rights reserved.
Conflict of interest statement
The authors have no conflict of interest to report.
Figures
Similar articles
-
Identification of a novel immune-related microRNA prognostic model in clear cell renal cell carcinoma.Transl Androl Urol. 2021 Feb;10(2):888-899. doi: 10.21037/tau-20-1495. Transl Androl Urol. 2021. PMID: 33718090 Free PMC article.
-
miR-19a correlates with poor prognosis of clear cell renal cell carcinoma patients via promoting cell proliferation and suppressing PTEN/SMAD4 expression.Int J Oncol. 2016 Dec;49(6):2589-2599. doi: 10.3892/ijo.2016.3746. Epub 2016 Oct 24. Int J Oncol. 2016. PMID: 27779660
-
Long Non-Coding RNA LUCAT1 Promotes Proliferation and Invasion in Clear Cell Renal Cell Carcinoma Through AKT/GSK-3β Signaling Pathway.Cell Physiol Biochem. 2018;48(3):891-904. doi: 10.1159/000491957. Epub 2018 Jul 20. Cell Physiol Biochem. 2018. PMID: 30032137
-
Molecular Mechanisms in Clear Cell Renal Cell Carcinoma: Role of miRNAs and Hypermethylated miRNA Genes in Crucial Oncogenic Pathways and Processes.Front Genet. 2019 Apr 24;10:320. doi: 10.3389/fgene.2019.00320. eCollection 2019. Front Genet. 2019. PMID: 31110513 Free PMC article. Review.
-
Systematic Analysis of microRNA Biomarkers for Diagnosis, Prognosis, and Therapy in Patients With Clear Cell Renal Cell Carcinoma.Front Oncol. 2020 Dec 4;10:543817. doi: 10.3389/fonc.2020.543817. eCollection 2020. Front Oncol. 2020. PMID: 33344224 Free PMC article.
Cited by
-
A "Lymphocyte MicroRNA Signature" as Predictive Biomarker of Immunotherapy Response and Plasma PD-1/PD-L1 Expression Levels in Patients with Metastatic Renal Cell Carcinoma: Pointing towards Epigenetic Reprogramming.Cancers (Basel). 2020 Nov 16;12(11):3396. doi: 10.3390/cancers12113396. Cancers (Basel). 2020. PMID: 33207823 Free PMC article.
-
MicroRNA Signature in Renal Cell Carcinoma.Front Oncol. 2020 Nov 30;10:596359. doi: 10.3389/fonc.2020.596359. eCollection 2020. Front Oncol. 2020. PMID: 33330087 Free PMC article. Review.
References
-
- Siegel RL, Miller KD, Jemal A. Cancer statistics, 2018. CA Cancer J Clin. 2018;68(1):7–30. - PubMed
-
- Ravaud A, Motzer RJ, Pandha HS, George DJ, Pantuck AJ, Patel A, et al. Adjuvant Sunitinib in High-Risk Renal-Cell Carcinoma after Nephrectomy. N Engl J Med. 2016;375(23):2246–54. - PubMed
-
- Frank I, Blute ML, Cheville JC, Lohse CM, Weaver AL, Leibovich BC, et al. A multifactorial postoperative surveillance model for patients with surgically treated clear cell renal cell carcinoma. J Urol. 2003;170(6 Pt 1):2225–32. - PubMed
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
