Differential skewing of donor-unrestricted and γδ T cell repertoires in tuberculosis-infected human lungs
- PMID: 31763997
- PMCID: PMC6934215
- DOI: 10.1172/JCI130711
Differential skewing of donor-unrestricted and γδ T cell repertoires in tuberculosis-infected human lungs
Abstract
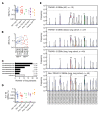
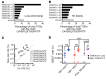
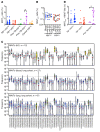
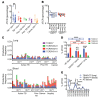
Unconventional T cells that recognize mycobacterial antigens are of great interest as potential vaccine targets against tuberculosis (TB). This includes donor-unrestricted T cells (DURTs), such as mucosa-associated invariant T cells (MAITs), CD1-restricted T cells, and γδ T cells. We exploited the distinctive nature of DURTs and γδ T cell receptors (TCRs) to investigate the involvement of these T cells during TB in the human lung by global TCR sequencing. Making use of surgical lung resections, we investigated the distribution, frequency, and characteristics of TCRs in lung tissue and matched blood from individuals infected with TB. Despite depletion of MAITs and certain CD1-restricted T cells from the blood, we found that the DURT repertoire was well preserved in the lungs, irrespective of disease status or HIV coinfection. The TCRδ repertoire, in contrast, was highly skewed in the lungs, where it was dominated by Vδ1 and distinguished by highly localized clonal expansions, consistent with the nonrecirculating lung-resident γδ T cell population. These data show that repertoire sequencing is a powerful tool for tracking T cell subsets during disease.
Keywords: Adaptive immunity; Immunology; Infectious disease; T-cell receptor; Tuberculosis.
Conflict of interest statement
Figures




Comment in
-
Clonal enrichments of Vδ2- γδ T cells in Mycobacterium tuberculosis-infected human lungs.J Clin Invest. 2020 Jan 2;130(1):68-70. doi: 10.1172/JCI133119. J Clin Invest. 2020. PMID: 31763996 Free PMC article.
References
-
- Global Tuberculosis Report 2017. World Health Organization. https://www.who.int/tb/publications/global_report/gtbr2017_main_text.pdf Accessed June 27, 2018.

