CRISPRitz: rapid, high-throughput and variant-aware in silico off-target site identification for CRISPR genome editing
- PMID: 31764961
- PMCID: PMC7141852
- DOI: 10.1093/bioinformatics/btz867
CRISPRitz: rapid, high-throughput and variant-aware in silico off-target site identification for CRISPR genome editing
Abstract
Motivation: Clustered regularly interspaced short palindromic repeats (CRISPR) technologies allow for facile genomic modification in a site-specific manner. A key step in this process is the in silico design of single guide RNAs to efficiently and specifically target a site of interest. To this end, it is necessary to enumerate all potential off-target sites within a given genome that could be inadvertently altered by nuclease-mediated cleavage. Currently available software for this task is limited by computational efficiency, variant support or annotation, and assessment of the functional impact of potential off-target effects.
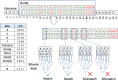
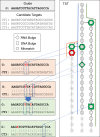
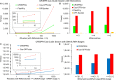
Results: To overcome these limitations, we have developed CRISPRitz, a suite of software tools to support the design and analysis of CRISPR/CRISPR-associated (Cas) experiments. Using efficient data structures combined with parallel computation, we offer a rapid, reliable, and exhaustive search mechanism to enumerate a comprehensive list of putative off-target sites. As proof-of-principle, we performed a head-to-head comparison with other available tools on several datasets. This analysis highlighted the unique features and superior computational performance of CRISPRitz including support for genomic searching with DNA/RNA bulges and mismatches of arbitrary size as specified by the user as well as consideration of genetic variants (variant-aware). In addition, graphical reports are offered for coding and non-coding regions that annotate the potential impact of putative off-target sites that lie within regions of functional genomic annotation (e.g. insulator and chromatin accessible sites from the ENCyclopedia Of DNA Elements [ENCODE] project).
Availability and implementation: The software is freely available at: https://github.com/pinellolab/CRISPRitzhttps://github.com/InfOmics/CRISPRitz.
Supplementary information: Supplementary data are available at Bioinformatics online.
© The Author(s) 2019. Published by Oxford University Press. All rights reserved. For permissions, please e-mail: journals.permissions@oup.com.
Figures



References
-
- Aho A.V., Corasick M.J. (1975) Efficient string matching: an aid to bibliographic search. Commun. ACM, 18, 333–340.
-
- Bentley J., Sedgewick B. (1998) Ternary search trees. Dr. Dobb’s J., 23,
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

