Complete Chloroplast Genomes of Vachellia nilotica and Senegalia senegal: Comparative Genomics and Phylogenomic Placement in a New Generic System
- PMID: 31765416
- PMCID: PMC6876885
- DOI: 10.1371/journal.pone.0225469
Complete Chloroplast Genomes of Vachellia nilotica and Senegalia senegal: Comparative Genomics and Phylogenomic Placement in a New Generic System
Abstract
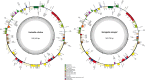
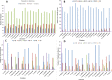
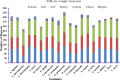
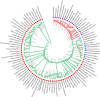
Vachellia and Senegalia are the most important genera in the subfamily Mimosoideae (Fabaceae). Recently, species from both genera were separated from the long-characterized Acacia due to their macro-morphological characteristics. However, this morpho-taxonomic differentiation struggles to discriminate some species, for example, Vachellia nilotica and Senegalia senegal. Therefore, sequencing the chloroplast (cp) genomes of these species and determining their phylogenetic placement via conserved genes may help to validate the taxonomy. Hence, we sequenced the cp genomes of V. nilotica and S. senegal, and the results showed that the sizes of the genomes are 165.3 and 162.7 kb, respectively. The cp genomes of both species comprised large single-copy regions (93,849~91,791 bp) and pairs of inverted repeats (IR; 26,093~26,008 bp). The total numbers of genes found in the V. nilotica and S. senegal cp genomes were 135 and 132, respectively. Approximately 123:130 repeats and 290:281 simple sequence repeats were found in the S. senegal and V. nilotica cp genomes, respectively. Genomic characterization was undertaken by comparing these genomes with those of 17 species belonging to related genera in Fabaceae. A phylogenetic analysis of the whole genome dataset and 56 shared genes was undertaken by generating cladograms with the same topologies and placing both species in a new generic system. These results support the likelihood of identifying segregate genera from Acacia with phylogenomic disposition of both V. nilotica and S. senegal in the subfamily Mimosoideae. The current study is the first to obtain complete genomic information on both species and may help to elucidate the genome architecture of these species and evaluate the genetic diversity among species.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures


Similar articles
-
The complete chloroplast genome sequences of Iris sanguinea donn ex Hornem.Mitochondrial DNA A DNA Mapp Seq Anal. 2017 Jan;28(1):15-16. doi: 10.3109/19401736.2015.1106521. Epub 2015 Dec 7. Mitochondrial DNA A DNA Mapp Seq Anal. 2017. PMID: 26641138
-
The complete chloroplast genome sequence of Maddenia hypoleuca koehne (Prunoideae, Rosaceae).Mitochondrial DNA A DNA Mapp Seq Anal. 2016 Nov;27(6):4651-4652. doi: 10.3109/19401736.2015.1106486. Epub 2015 Dec 28. Mitochondrial DNA A DNA Mapp Seq Anal. 2016. PMID: 26709445
-
Comparative Analysis of the Chloroplast Genomes of the Chinese Endemic Genus Urophysa and Their Contribution to Chloroplast Phylogeny and Adaptive Evolution.Int J Mol Sci. 2018 Jun 22;19(7):1847. doi: 10.3390/ijms19071847. Int J Mol Sci. 2018. PMID: 29932433 Free PMC article.
-
Rumors of Psychedelics, Psychotropics and Related Derivatives in Vachellia and Senegalia in Contrast with Verified Records in Australian Acacia.Plants (Basel). 2022 Dec 2;11(23):3356. doi: 10.3390/plants11233356. Plants (Basel). 2022. PMID: 36501395 Free PMC article. Review.
-
What is in a name? Scientific name changes of potentially poisonous plants and fungi in South Africa.J S Afr Vet Assoc. 2022 Nov;93(2):76-81. doi: 10.36303/JSAVA.160. Epub 2022 Jun 16. J S Afr Vet Assoc. 2022. PMID: 35934904 Review.
Cited by
-
Comparative Chloroplast Genomics of Endangered Euphorbia Species: Insights into Hotspot Divergence, Repetitive Sequence Variation, and Phylogeny.Plants (Basel). 2020 Feb 5;9(2):199. doi: 10.3390/plants9020199. Plants (Basel). 2020. PMID: 32033491 Free PMC article.
-
Complete Chloroplast Genomes of Acanthochlamys bracteata (China) and Xerophyta (Africa) (Velloziaceae): Comparative Genomics and Phylogenomic Placement.Front Plant Sci. 2021 Jun 14;12:691833. doi: 10.3389/fpls.2021.691833. eCollection 2021. Front Plant Sci. 2021. PMID: 34194461 Free PMC article.
-
Quantitative and Qualitative Genetic Studies of Some Acacia Species Grown in Egypt.Plants (Basel). 2020 Feb 13;9(2):243. doi: 10.3390/plants9020243. Plants (Basel). 2020. PMID: 32069993 Free PMC article.
-
Uncovering the first complete plastome genomics, comparative analyses, and phylogenetic dispositions of endemic medicinal plant Ziziphus hajarensis (Rhamnaceae).BMC Genomics. 2022 Jan 27;23(1):83. doi: 10.1186/s12864-022-08320-2. BMC Genomics. 2022. PMID: 35086490 Free PMC article.
-
Assembly, annotation and analysis of the chloroplast genome of the Algarrobo tree Neltuma pallida (subfamily: Caesalpinioideae).BMC Plant Biol. 2023 Nov 16;23(1):570. doi: 10.1186/s12870-023-04581-5. BMC Plant Biol. 2023. PMID: 37974117 Free PMC article.
References
-
- Haque A. Investigation of the fungi associated with dieback of prickly acacia (Vachellia nilotica subsp. indica) in Northern Australia. PhD, The University of Queensland. 2015.
-
- Kyalangalilwa B, Boatwright JS, Daru BH, Maurin O, van der Bank M. Phylogenetic position and revised classification of A cacia sl (F abaceae: M imosoideae) in A frica, including new combinations in V achellia and S enegalia. Botanical Journal of the Linnean Society. 2013;172(4):500–23.
-
- Beshai AA. The economics of a primary commodity: Gum Arabic. Oxford bulletin of economics and statistics. 1984;46(4):371–81.
-
- Khan IA, Abourashed EA. Leung's encyclopedia of common natural ingredients: used in food, drugs and cosmetics: John Wiley & Sons; 2011.
-
- Al-Assaf S, Phillips GO, Aoki H, Sasaki Y. Characterization and properties of Acacia senegal (L.) Willd. var. senegal with enhanced properties (Acacia (sen) SUPER GUM™): Part 1—Controlled maturation of Acacia senegal var. senegal to increase viscoelasticity, produce a hydrogel form and convert a poor into a good emulsifier. Food Hydrocolloids. 2007;21(3):319–28.
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Miscellaneous

