RITA Is Expressed in Trophoblastic Cells and Is Involved in Differentiation Processes of the Placenta
- PMID: 31766533
- PMCID: PMC6953008
- DOI: 10.3390/cells8121484
RITA Is Expressed in Trophoblastic Cells and Is Involved in Differentiation Processes of the Placenta
Abstract
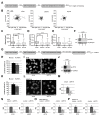
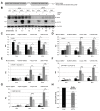
Preeclampsia (PE) remains a leading cause of maternal and perinatal mortality and morbidity worldwide. Its pathogenesis has not been fully elucidated and no causal therapy is currently available. It is of clinical relevance to decipher novel molecular biomarkers. RITA (RBP-J (recombination signal binding protein J)-interacting and tubulin-associated protein) has been identified as a negative modulator of the Notch pathway and as a microtubule-associated protein important for cell migration and invasion. In the present work, we have systematically studied RITA's expression in primary placental tissues from patients with early- and late-onset PE as well as in various trophoblastic cell lines. RITA is expressed in primary placental tissues throughout gestation, especially in proliferative villous cytotrophoblasts, in the terminally differentiated syncytiotrophoblast, and in migrating extravillous trophoblasts. RITA's messenger RNA (mRNA) level is decreased in primary tissue samples from early-onset PE patients. The deficiency of RITA impairs the motility and invasion capacity of trophoblastic cell lines, and compromises the fusion ability of trophoblast-derived choriocarcinoma cells. These data suggest that RITA may play important roles in the development of the placenta and possibly in the pathogenesis of PE.
Keywords: RITA; fusion; invasion; motility; preeclampsia; trophoblasts.
Conflict of interest statement
The authors declare no conflict of interest. The funders had no role in the study design; in collection, analyses, or interpretation of data; in manuscript writing, or in the decision to publish the results.
Figures




References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

