Whole Genome Sequencing of Aggregatibacter actinomycetemcomitans Cultured from Blood Stream Infections Reveals Three Major Phylogenetic Groups Including a Novel Lineage Expressing Serotype a Membrane O Polysaccharide
- PMID: 31766652
- PMCID: PMC6963875
- DOI: 10.3390/pathogens8040256
Whole Genome Sequencing of Aggregatibacter actinomycetemcomitans Cultured from Blood Stream Infections Reveals Three Major Phylogenetic Groups Including a Novel Lineage Expressing Serotype a Membrane O Polysaccharide
Abstract
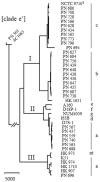
Twenty-nine strains of Aggregatibacter actinomycetemcomitans cultured from blood stream infections in Denmark were characterised. Serotyping was unremarkable, with almost equal proportions of the three major types plus a single serotype e strain. Whole genome sequencing positioned the serotype e strain outside the species boundary; moreover, one of the serotype a strains was unrelated to other strains of the major serotypes and to deposited sequences in the public databases. We identified five additional strains of this type in our collections. The particularity of the group was corroborated by phylogenetic analysis of concatenated core genes present in all strains of the species, and by uneven distribution of accessory genes only present in a subset of strains. Currently, the most accurate depiction of A. actinomycetemcomitans is a division into three lineages that differ in genomic content and competence for transformation. The clinical relevance of the different lineages is not known, and even strains excluded from the species sensu stricto can cause serious human infections. Serotyping is insufficient for characterisation, and serotypes a and e are not confined to specific lineages.
Keywords: average nucleotide identity; core and accessory genes; principal component analysis; taxonomy.
Conflict of interest statement
The authors declare no conflict of interest.
Figures


References
-
- Haubek D., Ennibi O.K., Poulsen K., Vaeth M., Poulsen S., Kilian M. Risk of aggressive periodontitis in adolescent carriers of the JP2 clone of Aggregatibacter (Actinobacillus) actinomycetemcomitans in Morocco: A prospective longitudinal cohort study. Lancet. 2008;371:237–242. doi: 10.1016/S0140-6736(08)60135-X. - DOI - PubMed
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

