DNA methylation aging clocks: challenges and recommendations
- PMID: 31767039
- PMCID: PMC6876109
- DOI: 10.1186/s13059-019-1824-y
DNA methylation aging clocks: challenges and recommendations
Abstract
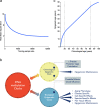
Epigenetic clocks comprise a set of CpG sites whose DNA methylation levels measure subject age. These clocks are acknowledged as a highly accurate molecular correlate of chronological age in humans and other vertebrates. Also, extensive research is aimed at their potential to quantify biological aging rates and test longevity or rejuvenating interventions. Here, we discuss key challenges to understand clock mechanisms and biomarker utility. This requires dissecting the drivers and regulators of age-related changes in single-cell, tissue- and disease-specific models, as well as exploring other epigenomic marks, longitudinal and diverse population studies, and non-human models. We also highlight important ethical issues in forensic age determination and predicting the trajectory of biological aging in an individual.
Conflict of interest statement
The authors declare that they have no competing interests, except for the following: WW is a co-founder of Cygenia GmbH (
Figures

References
-
- He W, Goodkind D, Kowal P. An aging world: 2015. 2016.
-
- WHO . The world report on ageing and health. 2015.
Publication types
MeSH terms
Grants and funding
- BB/PO28187/BB_/Biotechnology and Biological Sciences Research Council/United Kingdom
- BB/R00675X/1/BB_/Biotechnology and Biological Sciences Research Council/United Kingdom
- C18281/A19169/CRUK_/Cancer Research UK/United Kingdom
- BB/S020845/1/BB_/Biotechnology and Biological Sciences Research Council/United Kingdom
- U34 AG051425/AG/NIA NIH HHS/United States
- MC_UU_12013/5/MRC_/Medical Research Council/United Kingdom
- P30 ES009089/ES/NIEHS NIH HHS/United States
- CA207360/NH/NIH HHS/United States
- CA216265/NH/NIH HHS/United States
- AG021518/NH/NIH HHS/United States
- CA221705/NH/NIH HHS/United States
- R01 ES027747/NH/NIH HHS/United States
- U01 AG060908/AG/NIA NIH HHS/United States
- R01 ES025225/ES/NIEHS NIH HHS/United States
- 19169/CRUK_/Cancer Research UK/United Kingdom
- R01 ES025225/NH/NIH HHS/United States
- P01 AG047200/AG/NIA NIH HHS/United States
- MR/K013807/1/MRC_/Medical Research Council/United Kingdom
- P50 CA100632/CA/NCI NIH HHS/United States
- AG031862-12/AG/NIA NIH HHS/United States
- AG047745/NH/NIH HHS/United States
- MR/R005176/1/MRC_/Medical Research Council/United Kingdom
- R01 AG021518/AG/NIA NIH HHS/United States
- P01 AG031862/AG/NIA NIH HHS/United States
- R01 ES014811/ES/NIEHS NIH HHS/United States
- R01 CA080946/CA/NCI NIH HHS/United States
- R01 CA216265/CA/NCI NIH HHS/United States
- CA214005/NH/NIH HHS/United States
- R01 CA214005/CA/NCI NIH HHS/United States
- R01 CA207110/CA/NCI NIH HHS/United States
- CA080946/NH/NIH HHS/United States
- R01 AI121226/AI/NIAID NIH HHS/United States
- U54 CA221705/CA/NCI NIH HHS/United States
- RO1MH113930/NH/NIH HHS/United States
- R01 CA207360/CA/NCI NIH HHS/United States
- BB/K010867/1/BB_/Biotechnology and Biological Sciences Research Council/United Kingdom
- P30 ES009089/NH/NIH HHS/United States
- R01 ES027747/ES/NIEHS NIH HHS/United States
- DP1 AG047745/AG/NIA NIH HHS/United States
- BRC369/CN/SB/101310/DH_/Department of Health/United Kingdom
- CA207110/NH/NIH HHS/United States
- CA100632/NH/NIH HHS/United States
- RO1AI121226/NH/NIH HHS/United States
- MC_UU_00011/5/MRC_/Medical Research Council/United Kingdom
- R01ES014811/ES/NIEHS NIH HHS/United States
- R01 MH113930/MH/NIMH NIH HHS/United States
- U34AG051425-01/AG/NIA NIH HHS/United States
- RO1MDO1430401/NH/NIH HHS/United States
- AG047200/NH/NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

