Design of biochemical pattern forming systems from minimal motifs
- PMID: 31767054
- PMCID: PMC6922632
- DOI: 10.7554/eLife.48646
Design of biochemical pattern forming systems from minimal motifs
Abstract
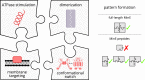
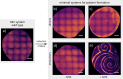
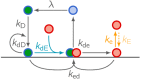
Although molecular self-organization and pattern formation are key features of life, only very few pattern-forming biochemical systems have been identified that can be reconstituted and studied in vitro under defined conditions. A systematic understanding of the underlying mechanisms is often hampered by multiple interactions, conformational flexibility and other complex features of the pattern forming proteins. Because of its compositional simplicity of only two proteins and a membrane, the MinDE system from Escherichia coli has in the past years been invaluable for deciphering the mechanisms of spatiotemporal self-organization in cells. Here, we explored the potential of reducing the complexity of this system even further, by identifying key functional motifs in the effector MinE that could be used to design pattern formation from scratch. In a combined approach of experiment and quantitative modeling, we show that starting from a minimal MinE-MinD interaction motif, pattern formation can be obtained by adding either dimerization or membrane-binding motifs. Moreover, we show that the pathways underlying pattern formation are recruitment-driven cytosolic cycling of MinE and recombination of membrane-bound MinE, and that these differ in their in vivo phenomenology.
Keywords: E. coli; in vitro reconstitution; min system; pattern formation; physics of living systems; reaction-diffusion; self-organization.
© 2019, Glock et al.
Conflict of interest statement
PG, FB, JH, EF, PS No competing interests declared
Figures







Similar articles
-
Min protein patterns emerge from rapid rebinding and membrane interaction of MinE.Nat Struct Mol Biol. 2011 May;18(5):577-83. doi: 10.1038/nsmb.2037. Epub 2011 Apr 24. Nat Struct Mol Biol. 2011. PMID: 21516096
-
Local Self-Enhancement of MinD Membrane Binding in Min Protein Pattern Formation.J Mol Biol. 2020 May 1;432(10):3191-3204. doi: 10.1016/j.jmb.2020.03.012. Epub 2020 Mar 19. J Mol Biol. 2020. PMID: 32199984
-
The E. coli MinCDE system in the regulation of protein patterns and gradients.Cell Mol Life Sci. 2019 Nov;76(21):4245-4273. doi: 10.1007/s00018-019-03218-x. Epub 2019 Jul 17. Cell Mol Life Sci. 2019. PMID: 31317204 Free PMC article. Review.
-
Direct MinE-membrane interaction contributes to the proper localization of MinDE in E. coli.Mol Microbiol. 2010 Jan;75(2):499-512. doi: 10.1111/j.1365-2958.2009.07006.x. Epub 2009 Dec 16. Mol Microbiol. 2010. PMID: 20025670 Free PMC article.
-
Spatial control of the cell division site by the Min system in Escherichia coli.Environ Microbiol. 2013 Dec;15(12):3229-39. doi: 10.1111/1462-2920.12119. Epub 2013 Apr 9. Environ Microbiol. 2013. PMID: 23574354 Review.
Cited by
-
Increasing MinD's Membrane Affinity Yields Standing Wave Oscillations and Functional Gradients on Flat Membranes.ACS Synth Biol. 2021 May 21;10(5):939-949. doi: 10.1021/acssynbio.0c00604. Epub 2021 Apr 21. ACS Synth Biol. 2021. PMID: 33881306 Free PMC article.
-
Noise-guided tuning of synthetic protein waves in living cells.bioRxiv [Preprint]. 2025 Mar 21:2025.03.21.644572. doi: 10.1101/2025.03.21.644572. bioRxiv. 2025. PMID: 40166177 Free PMC article. Preprint.
-
A Review of Mathematical and Computational Methods in Cancer Dynamics.Front Oncol. 2022 Jul 25;12:850731. doi: 10.3389/fonc.2022.850731. eCollection 2022. Front Oncol. 2022. PMID: 35957879 Free PMC article. Review.
-
Probing Biomolecular Interactions by a Pattern-Forming Peptide-Conjugate Sensor.Bioconjug Chem. 2021 Jan 20;32(1):172-181. doi: 10.1021/acs.bioconjchem.0c00596. Epub 2020 Dec 14. Bioconjug Chem. 2021. PMID: 33314917 Free PMC article.
-
ChemChaste: Simulating spatially inhomogeneous biochemical reaction-diffusion systems for modeling cell-environment feedbacks.Gigascience. 2022 Jun 17;11:giac051. doi: 10.1093/gigascience/giac051. Gigascience. 2022. PMID: 35715874 Free PMC article.
References
-
- Gessele R, Halatek J, Frey E. PAR protein activation-deactivation cycles stabilize long-axis polarization in c. elegans. bioRxiv. 2018 doi: 10.1101/451880. - DOI
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

