JASPer controls interphase histone H3S10 phosphorylation by chromosomal kinase JIL-1 in Drosophila
- PMID: 31767855
- PMCID: PMC6877644
- DOI: 10.1038/s41467-019-13174-6
JASPer controls interphase histone H3S10 phosphorylation by chromosomal kinase JIL-1 in Drosophila
Abstract
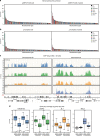
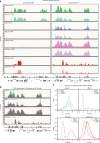
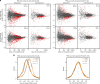
In flies, the chromosomal kinase JIL-1 is responsible for most interphase histone H3S10 phosphorylation and has been proposed to protect active chromatin from acquiring heterochromatic marks, such as dimethylated histone H3K9 (H3K9me2) and HP1. Here, we show that JIL-1's targeting to chromatin depends on a PWWP domain-containing protein JASPer (JIL-1 Anchoring and Stabilizing Protein). JASPer-JIL-1 (JJ)-complex is the major form of kinase in vivo and is targeted to active genes and telomeric transposons via binding of the PWWP domain of JASPer to H3K36me3 nucleosomes, to modulate transcriptional output. JIL-1 and JJ-complex depletion in cycling cells lead to small changes in H3K9me2 distribution at active genes and telomeric transposons. Finally, we identify interactors of the endogenous JJ-complex and propose that JIL-1 not only prevents heterochromatin formation but also coordinates chromatin-based regulation in the transcribed part of the genome.
Conflict of interest statement
The authors declare no competing interests.
Figures




References
-
- Adams RR, Maiato H, Earnshaw WC, Carmena M. Essential roles of Drosophila inner centromere protein (INCENP) and aurora B in histone H3 phosphorylation, metaphase chromosome alignment, kinetochore disjunction, and chromosome segregation. J. Cell Biol. 2001;153:865–880. doi: 10.1083/jcb.153.4.865. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials

