Designer DNA architecture offers precise and multivalent spatial pattern-recognition for viral sensing and inhibition
- PMID: 31767992
- PMCID: PMC6925649
- DOI: 10.1038/s41557-019-0369-8
Designer DNA architecture offers precise and multivalent spatial pattern-recognition for viral sensing and inhibition
Abstract
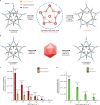
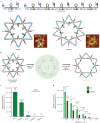
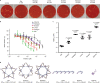
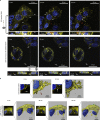
DNA, when folded into nanostructures with a specific shape, is capable of spacing and arranging binding sites into a complex geometric pattern with nanometre precision. Here we demonstrate a designer DNA nanostructure that can act as a template to display multiple binding motifs with precise spatial pattern-recognition properties, and that this approach can confer exceptional sensing and potent viral inhibitory capabilities. A star-shaped DNA architecture, carrying five molecular beacon-like motifs, was constructed to display ten dengue envelope protein domain III (ED3)-targeting aptamers into a two-dimensional pattern precisely matching the spatial arrangement of ED3 clusters on the dengue (DENV) viral surface. The resulting multivalent interactions provide high DENV-binding avidity. We show that this structure is a potent viral inhibitor and that it can act as a sensor by including a fluorescent output to report binding. Our molecular-platform design strategy could be adapted to detect and combat other disease-causing pathogens by generating the requisite ligand patterns on customized DNA nanoarchitectures.
Conflict of interest statement
The authors declare no competing interests.
Figures


References
-
- Harrison SC. Virology: looking inside adenovirus. Science. 2010;329:1026–1027. - PubMed
-
- Bandlow V, et al. Spatial screening of hemagglutinin on influenza A virus particles: sialyl-LacNAc displays on DNA and PEG scaffolds reveal the requirements for bivalency enhanced interactions with weak monovalent binders. J. Am. Chem. Soc. 2017;139:16389–16397. - PubMed
-
- Mourez M, et al. Designing a polyvalent inhibitor of anthrax toxin. Nat. Biotechnol. 2001;19:958–961. - PubMed
-
- Rai P, et al. Statistical pattern matching facilitates the design of polyvalent inhibitors of anthrax and cholera toxins. Nat. Biotechnol. 2006;24:582–586. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

