A Tppp3+Pdgfra+ tendon stem cell population contributes to regeneration and reveals a shared role for PDGF signalling in regeneration and fibrosis
- PMID: 31768046
- PMCID: PMC6895435
- DOI: 10.1038/s41556-019-0417-z
A Tppp3+Pdgfra+ tendon stem cell population contributes to regeneration and reveals a shared role for PDGF signalling in regeneration and fibrosis
Abstract
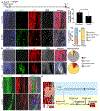
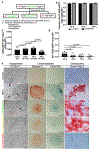
Tendon injuries cause prolonged disability and never recover completely. Current mechanistic understanding of tendon regeneration is limited. Here, we use single-cell transcriptomics to identify a tubulin polymerization-promoting protein family member 3-expressing (Tppp3+) cell population as potential tendon stem cells. Through inducible lineage tracing, we demonstrate that these cells can generate new tenocytes and self-renew upon injury. A fraction of Tppp3+ cells expresses platelet-derived growth factor receptor alpha (Pdfgra). Ectopic platelet-derived growth factor-AA (PDGF-AA) protein induces new tenocyte production while inactivation of Pdgfra in Tppp3+ cells blocks tendon regeneration. These results support Tppp3+Pdgfra+ cells as tendon stem cells. Unexpectedly, Tppp3-Pdgfra+ fibro-adipogenic progenitors coexist in the tendon stem cell niche and give rise to fibrotic cells, revealing a clandestine origin of fibrotic scars in healing tendons. Our results explain why fibrosis occurs in injured tendons and present clinical challenges to enhance tendon regeneration without a concurrent increase in fibrosis by PDGF application.
Conflict of interest statement
The authors declare no competing financial interests.
Competing Interests
The authors declare no conflicts of interest.
Figures

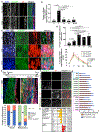

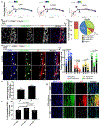



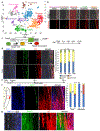



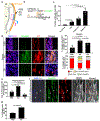

References
-
- Elliott DH. (1965) Structure and Function of Mammalian Tendon. Biol Rev Camb Philos Soc. 40:392–421. - PubMed
-
- Voleti PB, Buckley MR, Soslowsky LJ. (2012) Tendon Healing: Repair and Regeneration. Annu Rev Biomed Eng 14: 47–71. - PubMed
-
- Jozsa L and Kannus P. Human Tendons: Anatomy, Physiology and Pathology. Champaign: Human Kinetics, 1997. Print.
-
- Harvey T and Fan C-M (2018) Origins of tendon stem cells in situ. Front Biol 13(4): 263–276. DOI:10.1007/s11515-018-1504-4 - DOI
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Miscellaneous

