Diurnal oscillations of endogenous H2O2 sustained by p66Shc regulate circadian clocks
- PMID: 31768048
- PMCID: PMC7184292
- DOI: 10.1038/s41556-019-0420-4
Diurnal oscillations of endogenous H2O2 sustained by p66Shc regulate circadian clocks
Abstract
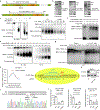
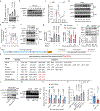
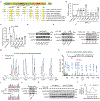
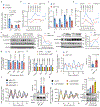
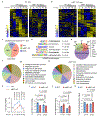
Redox balance, an essential feature of healthy physiological steady states, is regulated by circadian clocks, but whether or how endogenous redox signalling conversely regulates clockworks in mammals remains unknown. Here, we report circadian rhythms in the levels of endogenous H2O2 in mammalian cells and mouse livers. Using an unbiased method to screen for H2O2-sensitive transcription factors, we discovered that rhythmic redox control of CLOCK directly by endogenous H2O2 oscillations is required for proper intracellular clock function. Importantly, perturbations in the rhythm of H2O2 levels induced by the loss of p66Shc, which oscillates rhythmically in the liver and suprachiasmatic nucleus (SCN) of mice, disturb the rhythmic redox control of CLOCK function, reprogram hepatic transcriptome oscillations, lengthen the circadian period in mice and modulate light-induced clock resetting. Our findings suggest that redox signalling rhythms are intrinsically coupled to the circadian system through reversible oxidative modification of CLOCK and constitute essential mechanistic timekeeping components in mammals.
Conflict of interest statement
Competing interests
The authors declare no competing interests.
Figures




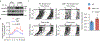




References
-
- Dodd AN et al. Plant circadian clocks increase photosynthesis, growth, survival, and competitive advantage. Science 309, 630–633 (2005). - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

