Exome sequencing in amyotrophic lateral sclerosis implicates a novel gene, DNAJC7, encoding a heat-shock protein
- PMID: 31768050
- PMCID: PMC6919277
- DOI: 10.1038/s41593-019-0530-0
Exome sequencing in amyotrophic lateral sclerosis implicates a novel gene, DNAJC7, encoding a heat-shock protein
Erratum in
-
Publisher Correction: Exome sequencing in amyotrophic lateral sclerosis implicates a novel gene, DNAJC7, encoding a heat-shock protein.Nat Neurosci. 2020 Feb;23(2):295. doi: 10.1038/s41593-019-0570-5. Nat Neurosci. 2020. PMID: 31857710
Abstract
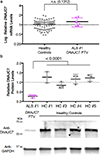
To discover novel genes underlying amyotrophic lateral sclerosis (ALS), we aggregated exomes from 3,864 cases and 7,839 ancestry-matched controls. We observed a significant excess of rare protein-truncating variants among ALS cases, and these variants were concentrated in constrained genes. Through gene level analyses, we replicated known ALS genes including SOD1, NEK1 and FUS. We also observed multiple distinct protein-truncating variants in a highly constrained gene, DNAJC7. The signal in DNAJC7 exceeded genome-wide significance, and immunoblotting assays showed depletion of DNAJC7 protein in fibroblasts in a patient with ALS carrying the p.Arg156Ter variant. DNAJC7 encodes a member of the heat-shock protein family, HSP40, which, along with HSP70 proteins, facilitates protein homeostasis, including folding of newly synthesized polypeptides and clearance of degraded proteins. When these processes are not regulated, misfolding and accumulation of aberrant proteins can occur and lead to protein aggregation, which is a pathological hallmark of neurodegeneration. Our results highlight DNAJC7 as a novel gene for ALS.
Conflict of interest statement
COMPETING INTERESTS
MN participation is supported by a consulting contract between Data Tecnica International and the National Institute on Aging, NIH, Bethesda, MD, USA, as a possible conflict of interest. MN also consults for Lysosomal Therapeutics Inc, the Michael J. Fox Foundation and Vivid Genomics among others. The other authors declare no competing interests.
Figures




References
Publication types
MeSH terms
Substances
Grants and funding
- U2C TR002818/TR/NCATS NIH HHS/United States
- MR/L501529/1/MRC_/Medical Research Council/United Kingdom
- U54 NS092091/NS/NINDS NIH HHS/United States
- G0600974/MRC_/Medical Research Council/United Kingdom
- G0500289/MRC_/Medical Research Council/United Kingdom
- SMITH/APR16/847-791/MNDA_/Motor Neurone Disease Association/United Kingdom
- G0900688/MRC_/Medical Research Council/United Kingdom
- MC_PC_17115/MRC_/Medical Research Council/United Kingdom
- U01 MH115727/MH/NIMH NIH HHS/United States
- G0900635/MRC_/Medical Research Council/United Kingdom
- ALCHALABI-DOBSON/APR14/829-791/MNDA_/Motor Neurone Disease Association/United Kingdom
- MR/L021803/1/MRC_/Medical Research Council/United Kingdom
- G1100695/MRC_/Medical Research Council/United Kingdom
- MR/R024804/1/MRC_/Medical Research Council/United Kingdom
- WT_/Wellcome Trust/United Kingdom
- MRF-060-0003-RG-SMITH/MRF_/MRF_/United Kingdom
- SHAW/NOV14/985-797/MNDA_/Motor Neurone Disease Association/United Kingdom
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

