Characterization and fine mapping of nonstop glumes 2 (nsg2) mutant in rice (Oryza sativa L.)
- PMID: 31768114
- PMCID: PMC6854344
- DOI: 10.5511/plantbiotechnology.19.0506a
Characterization and fine mapping of nonstop glumes 2 (nsg2) mutant in rice (Oryza sativa L.)
Abstract
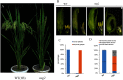
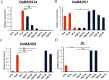
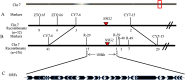
In cereal crops, the grain number per panicle and the grain yield are greatly affected by the number of florets in a spikelet. In wild-type rice, a spikelet only produces one fertile floret and beneath the floret are a pair of sterile lemmas and a pair of rudimentary glumes. This study characterized a rice spikelet mutant nonstop glumes 2 (nsg2). In the nsg2 mutant, both the sterile lemmas and rudimentary glumes were elongated, and part of sterile lemma looked like a lemma in appearance, shape and size. Detailed histological analysis and qPCR analysis revealed that the sterile lemmas in the nsg2 mutant had homeotically transformed into lemma-like organs. This phenotype indicates that NSG2 is involved in the regulation of spikelet development and supports the long-held view that sterile lemmas were derived from the lemmas of the two lateral florets. This implies that the rice spikelet has the potential to be restored to the "three florets spikelet", which may have existed in its ancestors. Genetic analysis reveals that the nsg2 trait is controlled by a single recessive gene. The NSG2 gene was finally mapped between markers R-20 and R-39 on chromosome 7 with a physical region of 180 kb. The two MYB family factors LOC_Os07g44030 and LOC_Os07g44090 might be involved in the development of the spikelet and floral organ, and they were considered as candidate genes of NSG2. These results provide a foundation for map-based cloning and function analysis of NSG2, as well as evidence to support "three-florets spikelet" breeding in rice.
Keywords: gene mapping; nonstop glumes 2 (nsg2); rice (Oryza sativa); spikelet.
© 2019 The Japanese Society for Plant Cell and Molecular Biology.
Figures


Similar articles
-
The pleiotropic SEPALLATA-like gene OsMADS34 reveals that the 'empty glumes' of rice (Oryza sativa) spikelets are in fact rudimentary lemmas.New Phytol. 2014 Apr;202(2):689-702. doi: 10.1111/nph.12657. Epub 2013 Dec 24. New Phytol. 2014. PMID: 24372518
-
LATERAL FLORET 2 encoding a sucrose non-fermenting 2 chromatin remodeling factor regulates axillary meristem of spikelet development in rice (Oryza sativa).New Phytol. 2025 Apr;246(2):598-615. doi: 10.1111/nph.20455. Epub 2025 Feb 25. New Phytol. 2025. PMID: 40007170
-
The rice pds1 locus genetically interacts with partner to cause panicle exsertion defects and ectopic tillers in spikelets.BMC Plant Biol. 2019 May 15;19(1):200. doi: 10.1186/s12870-019-1805-z. BMC Plant Biol. 2019. PMID: 31092192 Free PMC article.
-
Multifloret spikelet improves rice yield.New Phytol. 2020 Mar;225(6):2301-2306. doi: 10.1111/nph.16303. Epub 2019 Nov 29. New Phytol. 2020. PMID: 31677165 Review.
-
Of floral fortune: tinkering with the grain yield potential of cereal crops.New Phytol. 2020 Mar;225(5):1873-1882. doi: 10.1111/nph.16189. Epub 2019 Oct 11. New Phytol. 2020. PMID: 31509613 Review.
References
-
- Agrawal GK, Abe K, Yamazaki M, Miyao A, Hirochika H (2005) Conservation of the E-function for floral organ identity in rice revealed by the analysis of tissue culture-induced loss-of-function mutants of the OsMADS1 gene. Plant Mol Biol 59: 125–135 - PubMed
-
- Ashikari M, Sakakibara H, Lin SY, Yamamoto T, Takashi T, Nishimura A, Angeles ER, Qian Q, Kitano H, Matsuoka M (2005) Cytokinin oxidase regulates rice grain production. Science 309: 741–745 - PubMed
-
- Cusick F (1966) On phylogenetic and ontogenetic fusions. In: Gutter EG (ed) Trends in Plant Morphogenesis. Longmans, London, pp 170–183
LinkOut - more resources
Full Text Sources
