The role of noncoding mutations in blood cancers
- PMID: 31771951
- PMCID: PMC6899015
- DOI: 10.1242/dmm.041988
The role of noncoding mutations in blood cancers
Abstract
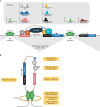
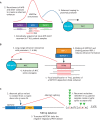
The search for oncogenic mutations in haematological malignancies has largely focused on coding sequence variants. These variants have been critical in understanding these complex cancers in greater detail, ultimately leading to better disease monitoring, subtyping and prognostication. In contrast, the search for oncogenic variants in the noncoding genome has proven to be challenging given the vastness of the search space, the intrinsic difficulty in assessing the impact of variants that do not code for functional proteins, and our still primitive understanding of the function harboured by large parts of the noncoding genome. Recent studies have broken ground on this quest, identifying somatically acquired and recurrent mutations in the noncoding genome that activate the expression of proto-oncogenes. In this Review, we explore some of the best-characterised examples of noncoding mutations in haematological malignancies, and highlight how a significant majority of these variants impinge on gene regulation through the formation of aberrant enhancers and promoters. We delve into the challenges faced by those that embark on a search for noncoding driver mutations, and provide a framework distilled from studies that have successfully identified such variants to overcome some of the most salient hurdles. Finally, we discuss the current therapeutic strategies being explored to target the oncogenic mechanism supported by recurrent noncoding variants. We postulate that the continued discovery and functional characterisation of somatic variants in the noncoding genome will not only advance our understanding of haematological malignancies, but offer novel therapeutic avenues and provide important insights into transcriptional regulation on a broader scale.
Keywords: Enhancers and promoters; Gene regulation; Haematological malignancy; Noncoding genome.
© 2019. Published by The Company of Biologists Ltd.
Conflict of interest statement
Competing interestsThe authors declare no competing or financial interests.
Figures

References
-
- Belver L., Yang A. Y., Albero R., Herranz D., Brundu F. G., Quinn S. A., Perez-Duran P., Alvarez S., Gianni F., Rashkovan M. et al. (2019). Gata3-controlled nucleosome eviction drives Myc enhancer activity in T-cell development and leukemia. Cancer Discov. 10.1158/2159-8290.CD-19-0471 - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

