Racial Differences in the Oral Microbiome: Data from Low-Income Populations of African Ancestry and European Ancestry
- PMID: 31771977
- PMCID: PMC6880044
- DOI: 10.1128/mSystems.00639-19
Racial Differences in the Oral Microbiome: Data from Low-Income Populations of African Ancestry and European Ancestry
Abstract
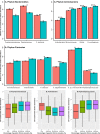
Increasing evidence indicates the significant racial difference in gut, vaginal, and skin microbiomes. However, little is known regarding the racial difference in the oral microbiome. In this study, deep sequencing of 16S rRNA genes was utilized to assess the oral microbiome in mouth rinse samples of 1,058 African-Americans (AAs) and 558 European-Americans (EAs) from the Southern Community Cohort Study. Generally, AAs had a higher species richness than EAs, with P = 5.28 × 10-14 (Wilcoxon rank sum test) for Faith's phylogenetic diversity index. A significant difference in overall microbiome composition was observed between AAs and EAs, with P = 5.94 × 10-4 (MiRKAT) for the weighted UniFrac distance matrix. We also found 32 bacterial taxa showing a significant differential abundance or prevalence between the two racial groups at a Bonferroni-corrected P < 0.05 in linear or logistic regression analyses. Generally, AAs showed a higher abundance of Bacteroidetes and a lower abundance of Actinobacteria and Firmicutes Interestingly, four periodontal pathogens, Porphyromonas gingivalis, Prevotella intermedia, Treponema denticola, and Filifactor alocis, were more prevalent among AAs than among EAs, with Bonferroni-corrected P values of 5.23 × 10-6, 4.47 × 10-6, 1.08 × 10-3, and 4.49 × 10-5, respectively. In addition, all of these 32 taxa were significantly correlated with the percentage of genetic African ancestry. These findings call for research to understand how the racial difference in oral microbiome influences the health disparity.IMPORTANCE In this systemic investigation of racial differences in the oral microbiome using a large data set, we disclosed the significant differences in the oral microbial richness/evenness, as well as in the overall microbial composition, between African-Americans and European-Americans. We also found multiple oral bacterial taxa, including several preidentified oral pathogens, showing a significant different abundance or prevalence between African-Americans and European-Americans. Furthermore, these taxa were consistently found to be associated with the percentage of genetic African ancestry. Our findings warrant further research to understand how the racial difference in the oral microbiome influences the health disparity.
Keywords: oral microbiome; racial difference.
Copyright © 2019 Yang et al.
Figures



References
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

