ILF3 is a substrate of SPOP for regulating serine biosynthesis in colorectal cancer
- PMID: 31772275
- PMCID: PMC7015059
- DOI: 10.1038/s41422-019-0257-1
ILF3 is a substrate of SPOP for regulating serine biosynthesis in colorectal cancer
Abstract
The Serine-Glycine-One-Carbon (SGOC) pathway is pivotal in multiple anabolic processes. Expression levels of SGOC genes are deregulated under tumorigenic conditions, suggesting participation of oncogenes in deregulating the SGOC biosynthetic pathway. However, the underlying mechanism remains elusive. Here, we identified that Interleukin enhancer-binding factor 3 (ILF3) is overexpressed in primary CRC patient specimens and correlates with poor prognosis. ILF3 is critical in regulating the SGOC pathway by directly regulating the mRNA stability of SGOC genes, thereby increasing SGOC genes expression and facilitating tumor growth. Mechanistic studies showed that the EGF-MEK-ERK pathway mediates ILF3 phosphorylation, which hinders E3 ligase speckle-type POZ protein (SPOP)-mediated poly-ubiquitination and degradation of ILF3. Significantly, combination of SGOC inhibitor and the anti-EGFR monoclonal antibody cetuximab can hinder the growth of patient-derived xenografts that sustain high ERK-ILF3 levels. Taken together, deregulation of ILF3 via the EGF-ERK signaling plays an important role in systemic serine metabolic reprogramming and confers a predilection toward CRC development. Our findings indicate that clinical evaluation of SGOC inhibitor is warranted for CRC patients with ILF3 overexpression.
Conflict of interest statement
The authors declare no competing interests.
Figures
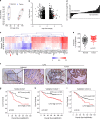


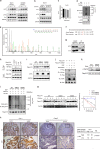



References
-
- Siegel RL, Miller KD, Jemal A. Cancer statistics, 2016. CA: Cancer J. Clin. 2016;66:7–30. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Research Materials
Miscellaneous

