Establishment of Best Practices for Evidence for Prediction: A Review
- PMID: 31774490
- PMCID: PMC7250718
- DOI: 10.1001/jamapsychiatry.2019.3671
Establishment of Best Practices for Evidence for Prediction: A Review
Abstract
Importance: Great interest exists in identifying methods to predict neuropsychiatric disease states and treatment outcomes from high-dimensional data, including neuroimaging and genomics data. The goal of this review is to highlight several potential problems that can arise in studies that aim to establish prediction.
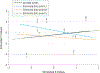
Observations: A number of neuroimaging studies have claimed to establish prediction while establishing only correlation, which is an inappropriate use of the statistical meaning of prediction. Statistical associations do not necessarily imply the ability to make predictions in a generalized manner; establishing evidence for prediction thus requires testing of the model on data separate from those used to estimate the model's parameters. This article discusses various measures of predictive performance and the limitations of some commonly used measures, with a focus on the importance of using multiple measures when assessing performance. For classification, the area under the receiver operating characteristic curve is an appropriate measure; for regression analysis, correlation should be avoided, and median absolute error is preferred.
Conclusions and relevance: To ensure accurate estimates of predictive validity, the recommended best practices for predictive modeling include the following: (1) in-sample model fit indices should not be reported as evidence for predictive accuracy, (2) the cross-validation procedure should encompass all operations applied to the data, (3) prediction analyses should not be performed with samples smaller than several hundred observations, (4) multiple measures of prediction accuracy should be examined and reported, (5) the coefficient of determination should be computed using the sums of squares formulation and not the correlation coefficient, and (6) k-fold cross-validation rather than leave-one-out cross-validation should be used.
Conflict of interest statement
Figures

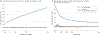
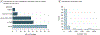
References
-
- Copas JB. Regression, prediction and shrinkage. J R Stat Soc Series B Stat Methodol. 1983;45(3):311354.

