Trans-omics Impact of Thymoproteasome in Cortical Thymic Epithelial Cells
- PMID: 31775054
- PMCID: PMC6897492
- DOI: 10.1016/j.celrep.2019.10.079
Trans-omics Impact of Thymoproteasome in Cortical Thymic Epithelial Cells
Abstract
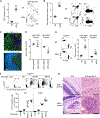
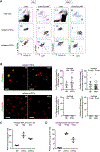
The thymic function to produce self-protective and self-tolerant T cells is chiefly mediated by cortical thymic epithelial cells (cTECs) and medullary TECs (mTECs). Recent studies including single-cell transcriptomic analyses have highlighted a rich diversity in functional mTEC subpopulations. Because of their limited cellularity, however, the biochemical characterization of TECs, including the proteomic profiling of cTECs and mTECs, has remained unestablished. Utilizing genetically modified mice that carry enlarged but functional thymuses, here we show a combination of proteomic and transcriptomic profiles for cTECs and mTECs, which identified signature molecules that characterize a developmental and functional contrast between cTECs and mTECs. Our results reveal a highly specific impact of the thymoproteasome on proteasome subunit composition in cTECs and provide an integrated trans-omics platform for further exploration of thymus biology.
Keywords: Psmb11; cortical thymic epithelial cells; cyclin D1; medullary thymic epithelial cells; proteome; thymoproteasome; trans-omics.
Published by Elsevier Inc.
Conflict of interest statement
DECLARATION OF INTERESTS
The authors declare no conflicting interests.
Figures





Comment in
-
Specific impact of β5t on proteasome subunit composition in cortical thymic epithelial cells.Cell Rep. 2021 Sep 7;36(10):109657. doi: 10.1016/j.celrep.2021.109657. Cell Rep. 2021. PMID: 34496235 Free PMC article.
-
PSMB11 regulates gene expression in cortical thymic epithelial cells.Cell Rep. 2021 Sep 7;36(10):109546. doi: 10.1016/j.celrep.2021.109546. Cell Rep. 2021. PMID: 34496243
References
-
- Anderson G, and Takahama Y (2012). Thymic epithelial cells: working class heroes for T cell development and repertoire selection. Trends Immunol. 33, 256–263. - PubMed
-
- Anderson G, Jenkinson EJ, Moore NC, and Owen JJ (1993). MHC class II-positive epithelium and mesenchyme cells are both required for T-cell development in the thymus. Nature 362, 70–73. - PubMed
-
- Anderson MS, Venanzi ES, Klein L, Chen Z, Berzins SP, Turley SJ, von Boehmer H, Bronson R, Dierich A, Benoist C, and Mathis D (2002). Projection of an immunological self shadow within the thymus by the aire protein. Science 298, 1395–1401. - PubMed
-
- Apavaloaei A, Brochu S, Dong M, Rouette A, Hardy MP, Villafano G, Murata S, Melichar HJ, and Perreault C (2019). PSMB11 orchestrates the development of CD4 and CD8 thymocytes via regulation of gene expression in cortical thymic epithelial cells. J. Immunol 202, 966–978. - PubMed
-
- Benjamini Y, and Hochberg Y (1995). Controlling the false discovery rate: a practical and powerful approach to multiple testing. J. R. Stat. Soc. B 57, 289–300.
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials

