A Comprehensive Analysis of Runs of Homozygosity of Eleven Cattle Breeds Representing Different Production Types
- PMID: 31775271
- PMCID: PMC6941163
- DOI: 10.3390/ani9121024
A Comprehensive Analysis of Runs of Homozygosity of Eleven Cattle Breeds Representing Different Production Types
Abstract
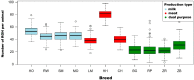
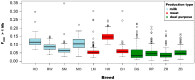
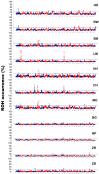
In the presented research, BovineSNP50 microarrays (Illumina) were applied to determine runs of homozygosity in the genomes of 11 cattle breeds maintained in Poland. These cattle breeds represent three basic utility types: milk, meat and dual purpose. Analysis of runs of homozygosity allowed the evaluation of the level of autozygosity within each breed in order to calculate the genomic inbreeding coefficient (FROH), as well as to identify regions of the genome with a high frequency of ROH occurrence, which may reflect traces of directional selectin left in their genomes. Visible differences in the length and distribution of runs of homozygosity in the genomes of the analyzed cattle breeds have been observed. The highest mean number and mean sums of lengths of runs of homozygosity were characteristic for Hereford cattle and intermediate for the Holstein-Friesian Black-and-White variety, Holstein-Friesian Red-and-White variety, Simmental, Limousin, Montbeliarde and Charolais breeds. However, lower values were observed for cattle of conserved breeds. Moreover, the selected livestock differed in the level of inbreeding estimated using the FROH coefficient. In regions of the genome with a high frequency of ROH occurrence, which may reflect the impact of directional selection, a number of genes were observed that can be potentially related to the production traits which are under selection pressure for specific production types. The most important detected genes were GHR, MSTN, DGAT1, FABP4, and TRH, with a known influence on the milk and meat traits of the studied cattle breeds.
Keywords: autozygosity; cattle; microarray; runs of homozygosity.
Conflict of interest statement
None of the authors have any relationships, financial or otherwise, with people or organizations that could have inappropriately influenced this work. The authors declare that they have no competing interests.
Figures


References
-
- Curik I., Ferenčaković M., Sölkner J. Inbreeding and runs of homozygosity: A possible solution to an old problem. Livest. Sci. 2014;166:26–34. doi: 10.1016/j.livsci.2014.05.034. - DOI
-
- Ferencakovic M., Hamzic E., Gredler B., Curik I., Sölkner J. Runs of homozygosity reveal genomewide autozygosity in the Austrian Fleckvieh cattle. Agric. Conspec. Sci. 2011;76:325–328.
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

