Transcriptome Analysis of Salt-Sensitive and Tolerant Genotypes Reveals Salt-Tolerance Metabolic Pathways in Sugar Beet
- PMID: 31775274
- PMCID: PMC6928841
- DOI: 10.3390/ijms20235910
Transcriptome Analysis of Salt-Sensitive and Tolerant Genotypes Reveals Salt-Tolerance Metabolic Pathways in Sugar Beet
Abstract
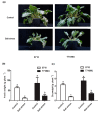
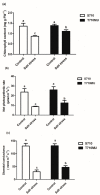
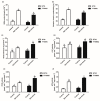
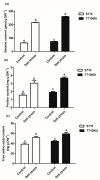
Soil salinization is a common environmental problem that seriously affects the yield and quality of crops. Sugar beet (Beta vulgaris L.), one of the main sugar crops in the world, shows a strong tolerance to salt stress. To decipher the molecular mechanism of sugar beet under salt stress, we conducted transcriptomic analyses of two contrasting sugar beet genotypes. To the best of our knowledge, this is the first comparison of salt-response transcriptomes in sugar beet with contrasting genotypes. Compared to the salt-sensitive cultivar (S710), the salt-tolerant one (T710MU) showed better growth and exhibited a higher chlorophyll content, higher antioxidant enzyme activity, and increased levels of osmotic adjustment molecules. Based on a high-throughput experimental system, 1714 differentially expressed genes were identified in the leaves of the salt-sensitive genotype, and 2912 in the salt-tolerant one. Many of the differentially expressed genes were involved in stress and defense responses, metabolic processes, signal transduction, transport processes, and cell wall synthesis. Moreover, expression patterns of several genes differed between the two cultivars in response to salt stress, and several key pathways involved in determining the salt tolerance of sugar beet, were identified. Our results revealed the mechanism of salt tolerance in sugar beet and provided potential metabolic pathways and gene markers for growing salt-tolerant cultivars.
Keywords: differentially expressed gene; physiological analysis; salt stress; sugar beet; transcriptomic analysis.
Conflict of interest statement
The authors declare no conflict of interest.
Figures



Similar articles
-
Comparative Physiological and Proteomic Analysis of Two Sugar Beet Genotypes with Contrasting Salt Tolerance.J Agric Food Chem. 2019 May 29;67(21):6056-6073. doi: 10.1021/acs.jafc.9b00244. Epub 2019 May 16. J Agric Food Chem. 2019. PMID: 31070911
-
De novo transcriptome assembly and identification of salt-responsive genes in sugar beet M14.Comput Biol Chem. 2018 Aug;75:1-10. doi: 10.1016/j.compbiolchem.2018.04.014. Epub 2018 Apr 22. Comput Biol Chem. 2018. PMID: 29705503
-
The physiological and metabolic changes in sugar beet seedlings under different levels of salt stress.J Plant Res. 2017 Nov;130(6):1079-1093. doi: 10.1007/s10265-017-0964-y. Epub 2017 Jul 15. J Plant Res. 2017. PMID: 28711996
-
Mechanisms of Sugar Beet Response to Biotic and Abiotic Stresses.Adv Exp Med Biol. 2020;1241:167-194. doi: 10.1007/978-3-030-41283-8_10. Adv Exp Med Biol. 2020. PMID: 32383121 Review.
-
Advances in Understanding the Physiological and Molecular Responses of Sugar Beet to Salt Stress.Front Plant Sci. 2019 Nov 6;10:1431. doi: 10.3389/fpls.2019.01431. eCollection 2019. Front Plant Sci. 2019. PMID: 31781145 Free PMC article. Review.
Cited by
-
The Roles of Calcineurin B-like Proteins in Plants under Salt Stress.Int J Mol Sci. 2023 Nov 30;24(23):16958. doi: 10.3390/ijms242316958. Int J Mol Sci. 2023. PMID: 38069281 Free PMC article. Review.
-
Transcriptomic and metabolomic analyses reveal mechanisms of adaptation to salinity in which carbon and nitrogen metabolism is altered in sugar beet roots.BMC Plant Biol. 2020 Apr 3;20(1):138. doi: 10.1186/s12870-020-02349-9. BMC Plant Biol. 2020. PMID: 32245415 Free PMC article.
-
Physiological and Proteomic Analysis of Different Molecular Mechanisms of Sugar Beet Response to Acidic and Alkaline pH Environment.Front Plant Sci. 2021 Jun 9;12:682799. doi: 10.3389/fpls.2021.682799. eCollection 2021. Front Plant Sci. 2021. PMID: 34178001 Free PMC article.
-
Combined transcriptomics and metabolomics analysis reveals salinity stress specific signaling and tolerance responses in the seagrass Zostera japonica.Plant Cell Rep. 2024 Jul 30;43(8):203. doi: 10.1007/s00299-024-03292-x. Plant Cell Rep. 2024. PMID: 39080075
-
Genomic and transcriptomic-based analysis of agronomic traits in sugar beet (Beta vulgaris L.) pure line IMA1.Front Plant Sci. 2022 Oct 13;13:1028885. doi: 10.3389/fpls.2022.1028885. eCollection 2022. Front Plant Sci. 2022. PMID: 36311117 Free PMC article.
References
-
- Abdel Latef A., He C. Effect of arbuscular mycorrhizal fungi on growth, mineral nutrition, antioxidant enzymes activity and fruit yield of tomato grown under salt stress. Sci. Hortic. 2011;127:228–233. doi: 10.1016/j.scienta.2010.09.020. - DOI
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

