Development of a high yielding expression platform for the introduction of non-natural amino acids in protein sequences
- PMID: 31775561
- PMCID: PMC6927762
- DOI: 10.1080/19420862.2019.1684749
Development of a high yielding expression platform for the introduction of non-natural amino acids in protein sequences
Abstract
The ability to genetically encode non-natural amino acids (nnAAs) into proteins offers an expanded tool set for protein engineering. nnAAs containing unique functional moieties have enabled the study of post-translational modifications, protein interactions, and protein folding. In addition, nnAAs have been developed that enable a variety of biorthogonal conjugation chemistries that allow precise and efficient protein conjugations. These are being studied to create the next generation of antibody-drug conjugates with improved efficacy, potency, and stability for the treatment of cancer. However, the efficiency of nnAA incorporation, and the productive yields of cell-based expression systems, have limited the utility and widespread use of this technology. We developed a process to isolate stable cell lines expressing a pyrrolysyl-tRNA synthetase/tRNApyl pair capable of efficient nnAA incorporation. Two different platform cell lines generated by these methods were used to produce IgG-expressing cell lines with normalized antibody titers of 3 g/L using continuous perfusion. We show that the antibodies produced by these platform cells contain the nnAA functionality that enables facile conjugations. Characterization of these highly active and robust platform hosts identified key parameters that affect nnAA incorporation efficiency. These highly efficient host platforms may help overcome the expression challenges that have impeded the developability of this technology for manufacturing proteins with nnAAs and represents an important step in expanding its utility.
Keywords: Antibody drug conjugate; IgG; PylRS; non-natural amino acid; tRNA.
Figures
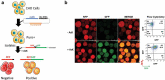
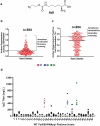



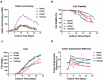


References
-
- Zhu GD, Fu YX. Design of next generation antibody drug conjugates. Yao Xue Xue Bao. Acta Pharmaceutica Sinica. 2013;48:1053–70. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
