The intestinal microbiota regulates host cholesterol homeostasis
- PMID: 31775890
- PMCID: PMC6882370
- DOI: 10.1186/s12915-019-0715-8
The intestinal microbiota regulates host cholesterol homeostasis
Abstract
Background: Management of blood cholesterol is a major focus of efforts to prevent cardiovascular diseases. The objective of this study was to investigate how the gut microbiota affects host cholesterol homeostasis at the organism scale.
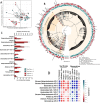
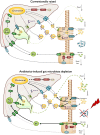
Results: We depleted the intestinal microbiota of hypercholesterolemic female Apoe-/- mice using broad-spectrum antibiotics. Measurement of plasma cholesterol levels as well as cholesterol synthesis and fluxes by complementary approaches showed that the intestinal microbiota strongly regulates plasma cholesterol level, hepatic cholesterol synthesis, and enterohepatic circulation. Moreover, transplant of the microbiota from humans harboring elevated plasma cholesterol levels to recipient mice induced a phenotype of high plasma cholesterol levels in association with a low hepatic cholesterol synthesis and high intestinal absorption pattern. Recipient mice phenotypes correlated with several specific bacterial phylotypes affiliated to Betaproteobacteria, Alistipes, Bacteroides, and Barnesiella taxa.
Conclusions: These results indicate that the intestinal microbiota determines the circulating cholesterol level and may thus represent a novel therapeutic target in the management of dyslipidemia and cardiovascular diseases.
Keywords: Antibiotics; Cholesterol metabolism; Dyslipidemia; Enterohepatic cycle; Intestinal microbiota; Microbiome; Microbiota transfer.
Conflict of interest statement
The authors declare that they have no competing interests.
Figures





References
-
- Vallejo-Vaz AJ, Robertson M, Catapano AL, Watts GF, Kastelein JJ, Packard CJ, et al. LDL-cholesterol lowering for the primary prevention of cardiovascular disease among men with primary elevations of LDL-cholesterol levels of 190 mg/dL or above: analyses from the WOSCOPS 5-year randomised trial and 20-year observational follow-up. Circulation. 2017;136(20):1878–1891. doi: 10.1161/CIRCULATIONAHA.117.027966. - DOI - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

