A novel faecal Lachnoclostridium marker for the non-invasive diagnosis of colorectal adenoma and cancer
- PMID: 31776231
- PMCID: PMC7306980
- DOI: 10.1136/gutjnl-2019-318532
A novel faecal Lachnoclostridium marker for the non-invasive diagnosis of colorectal adenoma and cancer
Abstract
Objective: There is a need for early detection of colorectal cancer (CRC) at precancerous-stage adenoma. Here, we identified novel faecal bacterial markers for diagnosing adenoma.
Design: This study included 1012 subjects (274 CRC, 353 adenoma and 385 controls) from two independent Asian groups. Candidate markers were identified by metagenomics and validated by targeted quantitative PCR.
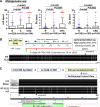
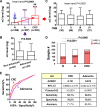
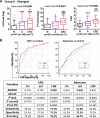
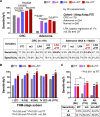
Results: Metagenomic analysis identified 'm3' from a Lachnoclostridium sp., Fusobacterium nucleatum (Fn) and Clostridium hathewayi (Ch) to be significantly enriched in adenoma. Faecal m3 and Fn were significantly increased from normal to adenoma to CRC (p<0.0001, linear trend by one-way ANOVA) in group I (n=698), which was further confirmed in group II (n=313; p<0.0001). Faecal m3 may perform better than Fn in distinguishing adenoma from controls (areas under the receiver operating characteristic curve (AUROCs) m3=0.675 vs Fn=0.620, p=0.09), while Fn performed better in diagnosing CRC (AUROCs Fn=0.862 vs m3=0.741, p<0.0001). At 78.5% specificity, m3 and Fn showed sensitivities of 48.3% and 33.8% for adenoma, and 62.1% and 77.8% for CRC, respectively. In a subgroup tested with faecal immunochemical test (FIT; n=642), m3 performed better than FIT in detecting adenoma (sensitivities for non-advanced and advanced adenomas of 44.2% and 50.8% by m3 (specificity=79.6%) vs 0% and 16.1% by FIT (specificity=98.5%)). Combining with FIT improved sensitivity of m3 for advanced adenoma to 56.8%. The combination of m3 with Fn, Ch, Bacteroides clarus and FIT performed best for diagnosing CRC (specificity=81.2% and sensitivity=93.8%).
Conclusion: This study identifies a novel bacterial marker m3 for the non-invasive diagnosis of colorectal adenoma.
Keywords: colonic bacteria; colorectal adenomas; colorectal cancer screening.
© Author(s) (or their employer(s)) 2020. Re-use permitted under CC BY-NC. No commercial re-use. See rights and permissions. Published by BMJ.
Conflict of interest statement
Competing interests: None declared.
Figures


References
-
- Allemani C, Matsuda T, Di Carlo V, et al. . Global surveillance of trends in cancer survival 2000–14 (CONCORD-3): analysis of individual records for 37 513 025 patients diagnosed with one of 18 cancers from 322 population-based registries in 71 countries. Lancet 2018;391:1023–75. 10.1016/S0140-6736(17)33326-3 - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Miscellaneous
