Combined HIV-1 sequence and integration site analysis informs viral dynamics and allows reconstruction of replicating viral ancestors
- PMID: 31776247
- PMCID: PMC6925994
- DOI: 10.1073/pnas.1910334116
Combined HIV-1 sequence and integration site analysis informs viral dynamics and allows reconstruction of replicating viral ancestors
Abstract
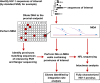
Understanding HIV-1 persistence despite antiretroviral therapy (ART) is of paramount importance. Both single-genome sequencing (SGS) and integration site analysis (ISA) provide useful information regarding the structure of persistent HIV DNA populations; however, until recently, there was no way to link integration sites to their cognate proviral sequences. Here, we used multiple-displacement amplification (MDA) of cellular DNA diluted to a proviral endpoint to obtain full-length proviral sequences and their corresponding sites of integration. We applied this method to lymph node and peripheral blood mononuclear cells from 5 ART-treated donors to determine whether groups of identical subgenomic sequences in the 2 compartments are the result of clonal expansion of infected cells or a viral genetic bottleneck. We found that identical proviral sequences can result from both cellular expansion and viral genetic bottlenecks occurring prior to ART initiation and following ART failure. We identified an expanded T cell clone carrying an intact provirus that matched a variant previously detected by viral outgrowth assays and expanded clones with wild-type and drug-resistant defective proviruses. We also found 2 clones from 1 donor that carried identical proviruses except for nonoverlapping deletions, from which we could infer the sequence of the intact parental virus. Thus, MDA-SGS can be used for "viral reconstruction" to better understand intrapatient HIV-1 evolution and to determine the clonality and structure of proviruses within expanded clones, including those with drug-resistant mutations. Importantly, we demonstrate that identical sequences observed by standard SGS are not always sufficient to establish proviral clonality.
Keywords: HIV persistence; integration site analysis; proviral structure.
Conflict of interest statement
Competing interest statement: J.W.M. is a consultant to Gilead Sciences, Merck Research Laboratories, Janssen Pharmaceuticals, and AccelevirDx, and a share option holder of Co-Crystal, Inc. B.F.K. and J.A.H. are co-authors on an October 2015 article. The remaining authors have no potential conflicts.
Figures




References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

