Genetic Diversity, Compartmentalization, and Age of HIV Proviruses Persisting in CD4+ T Cell Subsets during Long-Term Combination Antiretroviral Therapy
- PMID: 31776273
- PMCID: PMC7022348
- DOI: 10.1128/JVI.01786-19
Genetic Diversity, Compartmentalization, and Age of HIV Proviruses Persisting in CD4+ T Cell Subsets during Long-Term Combination Antiretroviral Therapy
Abstract
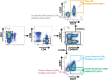
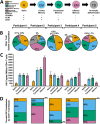
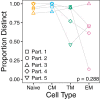
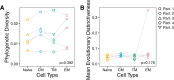
The HIV reservoir, which comprises diverse proviruses integrated into the genomes of infected, primarily CD4+ T cells, is the main barrier to developing an effective HIV cure. Our understanding of the genetics and dynamics of proviruses persisting within distinct CD4+ T cell subsets, however, remains incomplete. Using single-genome amplification, we characterized subgenomic proviral sequences (nef region) from naive, central memory, transitional memory, and effector memory CD4+ T cells from five HIV-infected individuals on long-term combination antiretroviral therapy (cART) and compared these to HIV RNA sequences isolated longitudinally from archived plasma collected prior to cART initiation, yielding HIV data sets spanning a median of 19.5 years (range, 10 to 20 years) per participant. We inferred a distribution of within-host phylogenies for each participant, from which we characterized proviral ages, phylogenetic diversity, and genetic compartmentalization between CD4+ T cell subsets. While three of five participants exhibited some degree of proviral compartmentalization between CD4+ T cell subsets, combined analyses revealed no evidence that any particular CD4+ T cell subset harbored the longest persisting, most genetically diverse, and/or most genetically distinctive HIV reservoir. In one participant, diverse proviruses archived within naive T cells were significantly younger than those in memory subsets, while for three other participants we observed no significant differences in proviral ages between subsets. In one participant, "old" proviruses were recovered from all subsets, and included one sequence, estimated to be 21.5 years old, that dominated (>93%) their effector memory subset. HIV eradication strategies will need to overcome within- and between-host genetic complexity of proviral landscapes, possibly via personalized approaches.IMPORTANCE The main barrier to HIV cure is the ability of a genetically diverse pool of proviruses, integrated into the genomes of infected CD4+ T cells, to persist despite long-term suppressive combination antiretroviral therapy (cART). CD4+ T cells, however, constitute a heterogeneous population due to their maturation across a developmental continuum, and the genetic "landscapes" of latent proviruses archived within them remains incompletely understood. We applied phylogenetic techniques, largely novel to HIV persistence research, to reconstruct within-host HIV evolutionary history and characterize proviral diversity in CD4+ T cell subsets in five individuals on long-term cART. Participants varied widely in terms of proviral burden, genetic diversity, and age distribution between CD4+ T cell subsets, revealing that proviral landscapes can differ between individuals and between infected cell types within an individual. Our findings expose each within-host latent reservoir as unique in its genetic complexity and support personalized strategies for HIV eradication.
Keywords: CD4+ T cells; HIV; cell subsets; cellular subsets; genetic compartmentalization; human immunodeficiency virus; persistence; proviral age; reservoir.
Copyright © 2020 American Society for Microbiology.
Figures






References
-
- Palella FJ Jr, Delaney KM, Moorman AC, Loveless MO, Fuhrer J, Satten GA, Aschman DJ, Holmberg SD. 1998. Declining morbidity and mortality among patients with advanced human immunodeficiency virus infection. HIV Outpatient Study Investigators. N Engl J Med 338:853–860. doi:10.1056/NEJM199803263381301. - DOI - PubMed
-
- Finzi D, Blankson J, Siliciano JD, Margolick JB, Chadwick K, Pierson T, Smith K, Lisziewicz J, Lori F, Flexner C, Quinn TC, Chaisson RE, Rosenberg E, Walker B, Gange S, Gallant J, Siliciano RF. 1999. Latent infection of CD4+ T cells provides a mechanism for lifelong persistence of HIV-1, even in patients on effective combination therapy. Nat Med 5:512–517. doi:10.1038/8394. - DOI - PubMed
-
- Finzi D, Hermankova M, Pierson T, Carruth LM, Buck C, Chaisson RE, Quinn TC, Chadwick K, Margolick J, Brookmeyer R, Gallant J, Markowitz M, Ho DD, Richman DD, Siliciano RF. 1997. Identification of a reservoir for HIV-1 in patients on highly active antiretroviral therapy. Science 278:1295–1300. doi:10.1126/science.278.5341.1295. - DOI - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials

