Nucleocapsid Protein Recruitment to Replication-Transcription Complexes Plays a Crucial Role in Coronaviral Life Cycle
- PMID: 31776274
- PMCID: PMC6997762
- DOI: 10.1128/JVI.01925-19
Nucleocapsid Protein Recruitment to Replication-Transcription Complexes Plays a Crucial Role in Coronaviral Life Cycle
Abstract
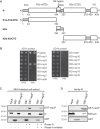
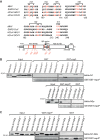
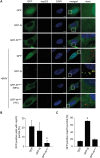
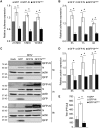
Coronavirus (CoV) nucleocapsid (N) proteins are key for incorporating genomic RNA into progeny viral particles. In infected cells, N proteins are present at the replication-transcription complexes (RTCs), the sites of CoV RNA synthesis. It has been shown that N proteins are important for viral replication and that the one of mouse hepatitis virus (MHV), a commonly used model CoV, interacts with nonstructural protein 3 (nsp3), a component of the RTCs. These two aspects of the CoV life cycle, however, have not been linked. We found that the MHV N protein binds exclusively to nsp3 and not other RTC components by using a systematic yeast two-hybrid approach, and we identified two distinct regions in the N protein that redundantly mediate this interaction. A selective N protein variant carrying point mutations in these two regions fails to bind nsp3 in vitro, resulting in inhibition of its recruitment to RTCs in vivo Furthermore, in contrast to the wild-type N protein, this N protein variant impairs the stimulation of genomic RNA and viral mRNA transcription in vivo and in vitro, which in turn leads to impairment of MHV replication and progeny production. Altogether, our results show that N protein recruitment to RTCs, via binding to nsp3, is an essential step in the CoV life cycle because it is critical for optimal viral RNA synthesis.IMPORTANCE CoVs have long been regarded as relatively harmless pathogens for humans. Severe respiratory tract infection outbreaks caused by severe acute respiratory syndrome CoV and Middle East respiratory syndrome CoV, however, have caused high pathogenicity and mortality rates in humans. These outbreaks highlighted the relevance of being able to control CoV infections. We used a model CoV, MHV, to investigate the importance of the recruitment of N protein, a central component of CoV virions, to intracellular platforms where CoVs replicate, transcribe, and translate their genomes. By identifying the principal binding partner at these intracellular platforms and generating a specific mutant, we found that N protein recruitment to these locations is crucial for promoting viral RNA synthesis. Moreover, blocking this recruitment strongly inhibits viral infection. Thus, our results explain an important aspect of the CoV life cycle and reveal an interaction of viral proteins that could be targeted in antiviral therapies.
Keywords: coronavirus; nucleocapsid N protein; replication-transcription complexes; viral mRNA synthesis.
Copyright © 2020 American Society for Microbiology.
Figures



References
-
- Woo PC, Lau SK, Lam CS, Lau CC, Tsang AK, Lau JH, Bai R, Teng JL, Tsang CC, Wang M, Zheng BJ, Chan KH, Yuen KY. 2012. Discovery of seven novel mammalian and avian coronaviruses in the genus Deltacoronavirus supports bat coronaviruses as the gene source of Alphacoronavirus and Betacoronavirus and avian coronaviruses as the gene source of Gammacoronavirus and Deltacoronavirus. J Virol 86:3995–4008. doi:10.1128/JVI.06540-11. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

