Boosting with AIDSVAX B/E Enhances Env Constant Region 1 and 2 Antibody-Dependent Cellular Cytotoxicity Breadth and Potency
- PMID: 31776278
- PMCID: PMC6997759
- DOI: 10.1128/JVI.01120-19
Boosting with AIDSVAX B/E Enhances Env Constant Region 1 and 2 Antibody-Dependent Cellular Cytotoxicity Breadth and Potency
Abstract
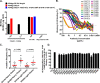
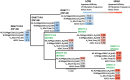
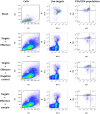
Induction of protective antibodies is a critical goal of HIV-1 vaccine development. One strategy is to induce nonneutralizing antibodies (NNAbs) that kill virus-infected cells, as these antibody specificities have been implicated in slowing HIV-1 disease progression and in protection. HIV-1 Env constant region 1 and 2 (C1C2) monoclonal antibodies (MAbs) frequently mediate potent antibody-dependent cellular cytotoxicity (ADCC), making them an important vaccine target. Here, we explore the effect of delayed and repetitive boosting of RV144 vaccine recipients with AIDSVAX B/E on the C1C2-specific MAb repertoire. It was found that boosting increased clonal lineage-specific ADCC breadth and potency. A ligand crystal structure of a vaccine-induced broad and potent ADCC-mediating C1C2-specific MAb showed that it bound a highly conserved Env gp120 epitope. Thus, boosting to affinity mature these types of IgG C1C2-specific antibody responses may be one method by which to make an improved HIV vaccine with higher efficacy than that seen in the RV144 trial.IMPORTANCE Over one million people become infected with HIV-1 each year, making the development of an efficacious HIV-1 vaccine an important unmet medical need. The RV144 human HIV-1 vaccine regimen is the only HIV-1 clinical trial to date to demonstrate vaccine efficacy. An area of focus has been on identifying ways by which to improve upon RV144 vaccine efficacy. The RV305 HIV-1 vaccine regimen was a follow-up boost of RV144 vaccine recipients that occurred 6 to 8 years after the conclusion of RV144. Our study focused on the effect of delayed boosting in humans on the vaccine-induced Env constant region 1 and 2 (C1C2)-specific antibody repertoire. It was found that boosting with an HIV-1 Env vaccine increased C1C2-specific antibody-dependent cellular cytotoxicity potency and breadth.
Keywords: HIV vaccine; antibody function.
Copyright © 2020 Easterhoff et al.
Figures
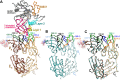




Similar articles
-
Recognition Patterns of the C1/C2 Epitopes Involved in Fc-Mediated Response in HIV-1 Natural Infection and the RV114 Vaccine Trial.mBio. 2020 Jun 30;11(3):e00208-20. doi: 10.1128/mBio.00208-20. mBio. 2020. PMID: 32605979 Free PMC article.
-
A Trimeric HIV-1 Envelope gp120 Immunogen Induces Potent and Broad Anti-V1V2 Loop Antibodies against HIV-1 in Rabbits and Rhesus Macaques.J Virol. 2018 Feb 12;92(5):e01796-17. doi: 10.1128/JVI.01796-17. Print 2018 Mar 1. J Virol. 2018. PMID: 29237847 Free PMC article.
-
Boosting of HIV envelope CD4 binding site antibodies with long variable heavy third complementarity determining region in the randomized double blind RV305 HIV-1 vaccine trial.PLoS Pathog. 2017 Feb 24;13(2):e1006182. doi: 10.1371/journal.ppat.1006182. eCollection 2017 Feb. PLoS Pathog. 2017. PMID: 28235027 Free PMC article. Clinical Trial.
-
Epitope specificity of human immunodeficiency virus-1 antibody dependent cellular cytotoxicity [ADCC] responses.Curr HIV Res. 2013 Jul;11(5):378-87. doi: 10.2174/1570162x113116660059. Curr HIV Res. 2013. PMID: 24191939 Free PMC article. Review.
-
Lessons from the RV144 Thai phase III HIV-1 vaccine trial and the search for correlates of protection.Annu Rev Med. 2015;66:423-37. doi: 10.1146/annurev-med-052912-123749. Epub 2014 Oct 17. Annu Rev Med. 2015. PMID: 25341006 Review.
Cited by
-
CD4 downregulation precedes Env expression and protects HIV-1-infected cells from ADCC mediated by non-neutralizing antibodies.mBio. 2024 Nov 13;15(11):e0182724. doi: 10.1128/mbio.01827-24. Epub 2024 Oct 7. mBio. 2024. PMID: 39373535 Free PMC article.
-
Recognition Patterns of the C1/C2 Epitopes Involved in Fc-Mediated Response in HIV-1 Natural Infection and the RV114 Vaccine Trial.mBio. 2020 Jun 30;11(3):e00208-20. doi: 10.1128/mBio.00208-20. mBio. 2020. PMID: 32605979 Free PMC article.
-
Effects of gp120 Inner Domain (ID2) Immunogen Doses on Elicitation of Anti-HIV-1 Functional Fc-Effector Response to C1/C2 (Cluster A) Epitopes in Mice.Microorganisms. 2020 Sep 28;8(10):1490. doi: 10.3390/microorganisms8101490. Microorganisms. 2020. PMID: 32998443 Free PMC article.
-
Improved killing of HIV-infected cells using three neutralizing and non-neutralizing antibodies.J Clin Invest. 2020 Oct 1;130(10):5157-5170. doi: 10.1172/JCI135557. J Clin Invest. 2020. PMID: 32584790 Free PMC article.
-
Viral vector delivered immunogen focuses HIV-1 antibody specificity and increases durability of the circulating antibody recall response.PLoS Pathog. 2023 May 31;19(5):e1011359. doi: 10.1371/journal.ppat.1011359. eCollection 2023 May. PLoS Pathog. 2023. PMID: 37256916 Free PMC article.
References
-
- Guan Y, Pazgier M, Sajadi MM, Kamin-Lewis R, Al-Darmarki S, Flinko R, Lovo E, Wu X, Robinson JE, Seaman MS, Fouts TR, Gallo RC, DeVico AL, Lewis GK. 2013. Diverse specificity and effector function among human antibodies to HIV-1 envelope glycoprotein epitopes exposed by CD4 binding. Proc Natl Acad Sci U S A 110:E69–E78. doi:10.1073/pnas.1217609110. - DOI - PMC - PubMed
-
- Ferrari G, Pollara J, Kozink D, Harms T, Drinker M, Freel S, Moody MA, Alam SM, Tomaras GD, Ochsenbauer C, Kappes JC, Shaw GM, Hoxie JA, Robinson JE, Haynes BF. 2011. An HIV-1 gp120 envelope human monoclonal antibody that recognizes a C1 conformational epitope mediates potent antibody-dependent cellular cytotoxicity (ADCC) activity and defines a common ADCC epitope in human HIV-1 serum. J Virol 85:7029–7036. doi:10.1128/JVI.00171-11. - DOI - PMC - PubMed
-
- Acharya P, Tolbert WD, Gohain N, Wu X, Yu L, Liu T, Huang W, Huang CC, Kwon YD, Louder RK, Luongo TS, McLellan JS, Pancera M, Yang Y, Zhang B, Flinko R, Foulke JS Jr, Sajadi MM, Kamin-Lewis R, Robinson JE, Martin L, Kwong PD, Guan Y, DeVico AL, Lewis GK, Pazgier M. 2014. Structural definition of an antibody-dependent cellular cytotoxicity response implicated in reduced risk for HIV-1 infection. J Virol 88:12895–12906. doi:10.1128/JVI.02194-14. - DOI - PMC - PubMed
-
- Tolbert WD, Gohain N, Veillette M, Chapleau JP, Orlandi C, Visciano ML, Ebadi M, DeVico AL, Fouts TR, Finzi A, Lewis GK, Pazgier M. 2016. Paring down HIV Env: design and crystal structure of a stabilized inner domain of HIV-1 gp120 displaying a major ADCC target of the A32 region. Structure 24:697–709. doi:10.1016/j.str.2016.03.005. - DOI - PMC - PubMed
-
- Gohain N, Tolbert WD, Acharya P, Yu L, Liu T, Zhao P, Orlandi C, Visciano ML, Kamin-Lewis R, Sajadi MM, Martin L, Robinson JE, Kwong PD, DeVico AL, Ray K, Lewis GK, Pazgier M. 2015. Cocrystal structures of antibody N60-i3 and antibody JR4 in complex with gp120 define more cluster A epitopes involved in effective antibody-dependent effector function against HIV-1. J Virol 89:8840–8854. doi:10.1128/JVI.01232-15. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

