A diarylamine derived from anthranilic acid inhibits ZIKV replication
- PMID: 31776405
- PMCID: PMC6881388
- DOI: 10.1038/s41598-019-54169-z
A diarylamine derived from anthranilic acid inhibits ZIKV replication
Abstract
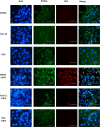
Zika virus (ZIKV) is a mosquito-transmitted Flavivirus, originally identified in Uganda in 1947 and recently associated with a large outbreak in South America. Despite extensive efforts there are currently no approved antiviral compounds for treatment of ZIKV infection. Here we describe the antiviral activity of diarylamines derived from anthranilic acid (FAMs) against ZIKV. A synthetic FAM (E3) demonstrated anti-ZIKV potential by reducing viral replication up to 86%. We analyzed the possible mechanisms of action of FAM E3 by evaluating the intercalation of this compound into the viral dsRNA and its interaction with the RNA polymerase of bacteriophage SP6. However, FAM E3 did not act by these mechanisms. In silico results predicted that FAM E3 might bind to the ZIKV NS3 helicase suggesting that this protein could be one possible target of this compound. To test this, the thermal stability and the ATPase activity of the ZIKV NS3 helicase domain (NS3Hel) were investigated in vitro and we demonstrated that FAM E3 could indeed bind to and stabilize NS3Hel.
Conflict of interest statement
The authors declare no competing interests.
Figures






References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical

