Genome size and endopolyploidy evolution across the moss phylogeny
- PMID: 31777923
- PMCID: PMC7102977
- DOI: 10.1093/aob/mcz194
Genome size and endopolyploidy evolution across the moss phylogeny
Abstract
Background and aims: Compared with other plant lineages, bryophytes have very small genomes with little variation across species, and high levels of endopolyploid nuclei. This study is the first analysis of moss genome evolution over a broad taxonomic sampling using phylogenetic comparative methods. We aim to determine whether genome size evolution is unidirectional as well as examine whether genome size and endopolyploidy are correlated in mosses.
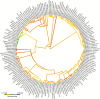
Methods: Genome size and endoreduplication index (EI) estimates were newly generated using flow cytometry from moss samples collected in Canada. Phylogenetic relationships between moss species were reconstructed using GenBank sequence data and maximum likelihood methods. Additional 1C-values were compiled from the literature and genome size and EI were mapped onto the phylogeny to reconstruct ancestral character states, test for phylogenetic signal and perform phylogenetic independent contrasts.
Key results: Genome size and EI were obtained for over 50 moss taxa. New genome size estimates are reported for 33 moss species and new EIs are reported for 20 species. In combination with data from the literature, genome sizes were mapped onto a phylogeny for 173 moss species with this analysis, indicating that genome size evolution in mosses does not appear to be unidirectional. Significant phylogenetic signal was detected for genome size when evaluated across the phylogeny, whereas phylogenetic signal was not detected for EI. Genome size and EI were not found to be significantly correlated when using phylogenetically corrected values.
Conclusions: Significant phylogenetic signal indicates closely related mosses have more similar genome sizes and EI values. This study supports that DNA content in mosses is defined by small genomes that are highly endopolyploid, suggesting strong selective pressure to maintain these features. Further research is needed to understand the functional significance of DNA content evolution in mosses.
Keywords: Bryophytes; endopolyploidy; endoreduplication; flow cytometry; genome size; mosses; phylogenetic independent contrasts (PIC); phylogenetic signal.
© The Author(s) 2019. Published by Oxford University Press on behalf of the Annals of Botany Company. All rights reserved. For permissions, please e-mail: journals.permissions@oup.com.
Figures


References
-
- Ågren JA, Wright SI. 2015. Selfish genetic elements and plant genome size evolution. Trends in Plant Science 20: 195–196. - PubMed
-
- Andrus RE. 1986. Some aspects of Sphagnum ecology. Canadian Journal of Botany 64: 416–426.
-
- Bainard JD, Newmaster SG. 2010. Endopolyploidy in bryophytes: widespread in mosses and absent in liverworts. Journal of Botany 2010: 316356.
-
- Bainard JD, Villarreal JC. 2013. Genome size increases in recently diverged hornwort clades. Genome 56: 431–435. - PubMed
-
- Bainard JD, Fazekas AJF, Newmaster SG. 2010. Methodology significantly affects genome size estimates: quantitative evidence using bryophytes. Cytometry Part A 77A: 725–732. - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources

