Characterization of zygotic genome activation-dependent maternal mRNA clearance in mouse
- PMID: 31777931
- PMCID: PMC6954448
- DOI: 10.1093/nar/gkz1111
Characterization of zygotic genome activation-dependent maternal mRNA clearance in mouse
Abstract
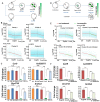
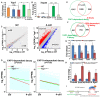
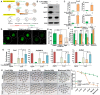
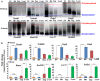
An important event of the maternal-to-zygotic transition (MZT) in animal embryos is the elimination of a subset of the maternal transcripts that accumulated during oogenesis. In both invertebrates and vertebrates, a maternally encoded mRNA decay pathway (M-decay) acts before zygotic genome activation (ZGA) while a second pathway, which requires zygotic transcription, subsequently clears additional mRNAs (Z-decay). To date the mechanisms that activate the Z-decay pathway in mammalian early embryos have not been investigated. Here, we identify murine maternal transcripts that are degraded after ZGA and show that inhibition of de novo transcription stabilizes these mRNAs in mouse embryos. We show that YAP1-TEAD4 transcription factor-mediated transcription is essential for Z-decay in mouse embryos and that TEAD4-triggered zygotic expression of terminal uridylyltransferases TUT4 and TUT7 and mRNA 3'-oligouridylation direct Z-decay. Components of the M-decay pathway, including BTG4 and the CCR4-NOT deadenylase, continue to function in Z-decay but require reinforcement from the zygotic factors for timely removal of maternal mRNAs. A long 3'-UTR and active translation confer resistance of Z-decay transcripts to M-decay during oocyte meiotic maturation. The Z-decay pathway is required for mouse embryo development beyond the four-cell stage and contributes to the developmental competence of preimplantation embryos.
© The Author(s) 2019. Published by Oxford University Press on behalf of Nucleic Acids Research.
Figures


References
-
- Sha Q.Q., Zhang J., Fan H.Y.. A story of birth and death: mRNA translation and clearance at the onset of maternal-to-zygotic transition in mammals. Biol. Reprod. 2019; 101:579–590. - PubMed
-
- Tadros W., Lipshitz H.D.. The maternal-to-zygotic transition: a play in two acts. Development. 2009; 136:3033–3042. - PubMed
-
- Walser C.B., Lipshitz H.D.. Transcript clearance during the maternal-to-zygotic transition. Curr. Opin. Genet. Dev. 2011; 21:431–443. - PubMed
-
- Bashirullah A., Halsell S.R., Cooperstock R.L., Kloc M., Karaiskakis A., Fisher W.W., Fu W., Hamilton J.K., Etkin L.D., Lipshitz H.D.. Joint action of two RNA degradation pathways controls the timing of maternal transcript elimination at the midblastula transition in Drosophila melanogaster. EMBO J. 1999; 18:2610–2620. - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

