ClinVar: improvements to accessing data
- PMID: 31777943
- PMCID: PMC6943040
- DOI: 10.1093/nar/gkz972
ClinVar: improvements to accessing data
Abstract
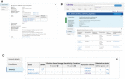
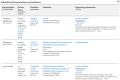
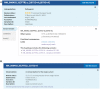
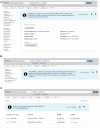
ClinVar is a freely available, public archive of human genetic variants and interpretations of their relationships to diseases and other conditions, maintained at the National Institutes of Health (NIH). Submitted interpretations of variants are aggregated and made available on the ClinVar website (https://www.ncbi.nlm.nih.gov/clinvar/), and as downloadable files via FTP and through programmatic tools such as NCBI's E-utilities. The default view on the ClinVar website, the Variation page, was recently redesigned. The new layout includes several new sections that make it easier to find submitted data as well as summary data such as all diseases and citations reported for the variant. The new design also better represents more complex data such as haplotypes and genotypes, as well as variants that are in ClinVar as part of a haplotype or genotype but have no interpretation for the single variant. ClinVar's variant-centric XML had its production release in April 2019. The ClinVar website and E-utilities both have been updated to support the VCV (variation in ClinVar) accession numbers found in the variant-centric XML file. ClinVar's search engine has been fine-tuned for improved retrieval of search results.
Published by Oxford University Press on behalf of Nucleic Acids Research 2019.
Figures


References
-
- Rubinstein W.S., Maglott D.R., Lee J.M., Kattman B.L., Malheiro A.J., Ovetsky M., Hem V., Gorelenkov V., Song G., Wallin C. et al. .. The NIH genetic testing registry: a new, centralized database of genetic tests to enable access to comprehensive information and improve transparency. Nucleic Acids Res. 2013; 41:D925–D935. - PMC - PubMed
-
- ACMG Board of Directors Laboratory and clinical genomic data sharing is crucial to improving genetic health care: a position statement of the American College of Medical Genetics and Genomics. Genet. Med. 2017; 19:721–722. - PubMed
-
- National Society of Genetic Counselors (NSGC) 2015; Clinical Data Sharing (Position Statement) [Blog post].
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

