TLR8 Is a Sensor of RNase T2 Degradation Products
- PMID: 31778653
- PMCID: PMC7116005
- DOI: 10.1016/j.cell.2019.11.001
TLR8 Is a Sensor of RNase T2 Degradation Products
Abstract
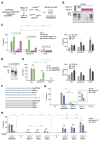
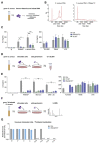
TLR8 is among the highest-expressed pattern-recognition receptors in the human myeloid compartment, yet its mode of action is poorly understood. TLR8 engages two distinct ligand binding sites to sense RNA degradation products, although it remains unclear how these ligands are formed in cellulo in the context of complex RNA molecule sensing. Here, we identified the lysosomal endoribonuclease RNase T2 as a non-redundant upstream component of TLR8-dependent RNA recognition. RNase T2 activity is required for rendering complex single-stranded, exogenous RNA molecules detectable for TLR8. This is due to RNase T2's preferential cleavage of single-stranded RNA molecules between purine and uridine residues, which critically contributes to the supply of catabolic uridine and the generation of purine-2',3'-cyclophosphate-terminated oligoribonucleotides. Thus-generated molecules constitute agonistic ligands for the first and second binding pocket of TLR8. Together, these results establish the identity and origin of the RNA-derived molecular pattern sensed by TLR8.
Keywords: RNA; RNase T2; TLR8; innate immunity; macrophage; monocyte; pattern recognition; toll-like receptor.
Copyright © 2019 Elsevier Inc. All rights reserved.
Conflict of interest statement
The authors declare no competing interests
Figures
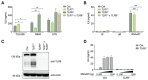
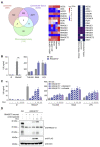
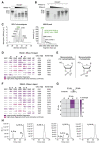
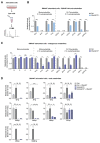

References
-
- Ablasser A, Poeck H, Anz D, Berger M, Schlee M, Kim S, Bourquin C, Goutagny N, Jiang Z, Fitzgerald KA, et al. Selection of molecular structure and delivery of RNA oligonucleotides to activate TLR7 versus TLR8 and to induce high amounts of IL-12p70 in primary human monocytes. J Immunol. 2009;182:6824–6833. - PubMed
-
- Barbalat R, Ewald SE, Mouchess ML, Barton GM. Nucleic acid recognition by the innate immune system. Annu Rev Immunol. 2011;29:185–214. - PubMed
-
- Bergstrom B, Aune MH, Awuh JA, Kojen JF, Blix KJ, Ryan L, Flo TH, Mollnes TE, Espevik T, Stenvik J. TLR8 Senses Staphylococcus aureus RNA in Human Primary Monocytes and Macrophages and Induces IFN-beta Production via a TAK1-IKKbeta-IRF5 Signaling Pathway. J Immunol. 2015;195:1100–1111. - PubMed
-
- Chan MP, Onji M, Fukui R, Kawane K, Shibata T, Saitoh S, Ohto U, Shimizu T, Barber GN, Miyake K. DNase II-dependent DNA digestion is required for DNA sensing by TLR9. Nat Commun. 2015;6 5853. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

