Insights into Innate Sensing of Prototype Foamy Viruses in Myeloid Cells
- PMID: 31779173
- PMCID: PMC6950106
- DOI: 10.3390/v11121095
Insights into Innate Sensing of Prototype Foamy Viruses in Myeloid Cells
Abstract
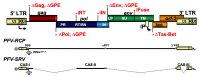
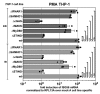
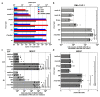
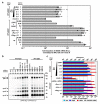
Foamy viruses (FVs) belong to the Spumaretrovirinae subfamily of retroviruses and are characterized by unique features in their replication strategy. This includes a reverse transcription (RTr) step of the packaged RNA genome late in replication, resulting in the release of particles with a fraction of them already containing an infectious viral DNA (vDNA) genome. Little is known about the immune responses against FVs in their hosts, which control infection and may be responsible for their apparent apathogenic nature. We studied the interaction of FVs with the innate immune system in myeloid cells, and characterized the viral pathogen-associated molecular patterns (PAMPs) and the cellular pattern recognition receptors and sensing pathways involved. Upon cytoplasmic access, full-length but not minimal vector genome containing FVs with active reverse transcriptase, induced an efficient innate immune response in various myeloid cells. It was dependent on cellular cGAS and STING and largely unaffected by RTr inhibition during viral entry. This suggests that RTr products, which are generated during FV morphogenesis in infected cells, and are therefore already present in FV particles taken up by immune cells, are the main PAMPs of FVs with full-length genomes sensed in a cGAS and STING-dependent manner by the innate immune system in host cells of the myeloid lineage.
Keywords: STING; cGAS; foamy virus; innate sensing; retrovirus; spumavirus.
Conflict of interest statement
The authors declare no conflict of interest. The funders had no role in the design of the study; in the collection, analyses, or interpretation of data; in the writing of the manuscript, or in the decision to publish the results.
Figures


References
-
- Rethwilm A., Lindemann D. Foamy viruses. In: Knipe D.M., Howley P.M., editors. Fields Virology. 6th ed. Volume 2. Lippincott Williams & Wilkins, a Wolters Kluwer business; Philadelphia, PA, USA: 2013. pp. 1613–1632.
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Research Materials

