GABAergic Restriction of Network Dynamics Regulates Interneuron Survival in the Developing Cortex
- PMID: 31780329
- PMCID: PMC6982374
- DOI: 10.1016/j.neuron.2019.10.008
GABAergic Restriction of Network Dynamics Regulates Interneuron Survival in the Developing Cortex
Abstract
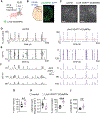
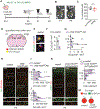
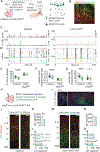
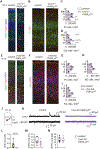
During neonatal development, sensory cortices generate spontaneous activity patterns shaped by both sensory experience and intrinsic influences. How these patterns contribute to the assembly of neuronal circuits is not clearly understood. Using longitudinal in vivo calcium imaging in un-anesthetized mouse pups, we show that spatially segregated functional assemblies composed of interneurons and pyramidal cells are prominent in the somatosensory cortex by postnatal day (P) 7. Both reduction of GABA release and synaptic inputs onto pyramidal cells erode the emergence of functional topography, leading to increased network synchrony. This aberrant pattern effectively blocks interneuron apoptosis, causing increased survival of parvalbumin and somatostatin interneurons. Furthermore, the effect of GABA on apoptosis is mediated by inputs from medial ganglionic eminence (MGE)-derived but not caudal ganglionic eminence (CGE)-derived interneurons. These findings indicate that immature MGE interneurons are fundamental for shaping GABA-driven activity patterns that balance the number of interneurons integrating into maturing cortical networks.
Keywords: GDPs; MGE; apoptosis; barrel cortex; calcium imaging; cell death; development; interneuron; spontaneous activity.
Copyright © 2019 Elsevier Inc. All rights reserved.
Conflict of interest statement
Declaration of Interests
The authors declare no competing interests.
Figures




References
-
- Anderson SA, Eisenstat DD, Shi L, and Rubenstein JL. 1997. ‘Interneuron migration from basal forebrain to neocortex: dependence on Dlx genes’, Science, 278: 474–6. - PubMed
-
- Arlotta P, Molyneaux BJ, Chen J, Inoue J, Kominami R, and Macklis JD. 2005. ‘Neuronal subtype-specific genes that control corticospinal motor neuron development in vivo’, Neuron, 45: 207–21. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials

