Accurate, scalable and integrative haplotype estimation
- PMID: 31780650
- PMCID: PMC6882857
- DOI: 10.1038/s41467-019-13225-y
Accurate, scalable and integrative haplotype estimation
Abstract
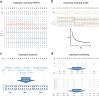
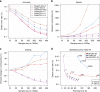
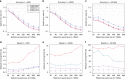
The number of human genomes being genotyped or sequenced increases exponentially and efficient haplotype estimation methods able to handle this amount of data are now required. Here we present a method, SHAPEIT4, which substantially improves upon other methods to process large genotype and high coverage sequencing datasets. It notably exhibits sub-linear running times with sample size, provides highly accurate haplotypes and allows integrating external phasing information such as large reference panels of haplotypes, collections of pre-phased variants and long sequencing reads. We provide SHAPEIT4 in an open source format and demonstrate its performance in terms of accuracy and running times on two gold standard datasets: the UK Biobank data and the Genome In A Bottle.
Conflict of interest statement
E.T.D. is chairman and member of the Board, Hybridstat Ltd. O.D., J.-F.Z., M.R.R., and J.L.M. declare no competing interests.
Figures


Similar articles
-
Haplotype estimation for biobank-scale data sets.Nat Genet. 2016 Jul;48(7):817-20. doi: 10.1038/ng.3583. Epub 2016 Jun 6. Nat Genet. 2016. PMID: 27270105 Free PMC article.
-
Fast two-stage phasing of large-scale sequence data.Am J Hum Genet. 2021 Oct 7;108(10):1880-1890. doi: 10.1016/j.ajhg.2021.08.005. Epub 2021 Sep 2. Am J Hum Genet. 2021. PMID: 34478634 Free PMC article.
-
Integrating read-based and population-based phasing for dense and accurate haplotyping of individual genomes.Bioinformatics. 2019 Jul 15;35(14):i242-i248. doi: 10.1093/bioinformatics/btz329. Bioinformatics. 2019. PMID: 31510646 Free PMC article.
-
trioPhaser: using Mendelian inheritance logic to improve genomic phasing of trios.BMC Bioinformatics. 2021 Nov 22;22(1):559. doi: 10.1186/s12859-021-04470-4. BMC Bioinformatics. 2021. PMID: 34809557 Free PMC article.
-
Haplotyping-Assisted Diploid Assembly and Variant Detection with Linked Reads.Methods Mol Biol. 2023;2590:161-182. doi: 10.1007/978-1-0716-2819-5_11. Methods Mol Biol. 2023. PMID: 36335499 Review.
Cited by
-
An update on the neurological short tandem repeat expansion disorders and the emergence of long-read sequencing diagnostics.Acta Neuropathol Commun. 2021 May 25;9(1):98. doi: 10.1186/s40478-021-01201-x. Acta Neuropathol Commun. 2021. PMID: 34034831 Free PMC article. Review.
-
Probabilistic Estimation of Identity by Descent Segment Endpoints and Detection of Recent Selection.Am J Hum Genet. 2020 Nov 5;107(5):895-910. doi: 10.1016/j.ajhg.2020.09.010. Epub 2020 Oct 13. Am J Hum Genet. 2020. PMID: 33053335 Free PMC article.
-
Genomic basis of the giga-chromosomes and giga-genome of tree peony Paeonia ostii.Nat Commun. 2022 Nov 28;13(1):7328. doi: 10.1038/s41467-022-35063-1. Nat Commun. 2022. PMID: 36443323 Free PMC article.
-
Precision medicine via the integration of phenotype-genotype information in neonatal genome project.Fundam Res. 2022 Jul 21;2(6):873-884. doi: 10.1016/j.fmre.2022.07.003. eCollection 2022 Nov. Fundam Res. 2022. PMID: 38933389 Free PMC article. Review.
-
De novo homozygous variant of the SCN1A gene in a patient with severe Dravet syndrome complicated by acute encephalopathy.Neurogenetics. 2021 May;22(2):133-136. doi: 10.1007/s10048-021-00636-7. Epub 2021 Mar 5. Neurogenetics. 2021. PMID: 33674996
References
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

