Rare Variants in 48 Genes Account for 42% of Cases of Epilepsy With or Without Neurodevelopmental Delay in 246 Pediatric Patients
- PMID: 31780880
- PMCID: PMC6856296
- DOI: 10.3389/fnins.2019.01135
Rare Variants in 48 Genes Account for 42% of Cases of Epilepsy With or Without Neurodevelopmental Delay in 246 Pediatric Patients
Abstract
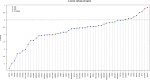
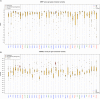
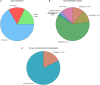
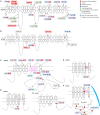
In order to characterize the genetic architecture of epilepsy in a pediatric population from the Iberian Peninsula (including the Canary Islands), we conducted targeted exome sequencing of 246 patients with infantile-onset seizures with or without neurodevelopmental delay. We detected 107 variants in 48 different genes, which were implicated in neuronal excitability, neurodevelopment, synaptic transmission, and metabolic pathways. In 104 cases (42%) we detected variant(s) that we classified as pathogenic or likely pathogenic. Of the 48 mutated genes, 32 were dominant, 8 recessive and 8 X-linked. Of the patients for whom family studies could be performed and in whom pathogenic variants were identified in dominant or X-linked genes, 82% carried de novo mutations. The involvement of small copy number variations (CNVs) is 9%. The use of progressively updated custom panels with high mean vertical coverage enabled establishment of a definitive diagnosis in a large proportion of cases (42%) and detection of CNVs (even duplications) with high fidelity. In 10.5% of patients we detected associations that are pending confirmation via functional and/or familial studies. Our findings had important consequences for the clinical management of the probands, since a large proportion of the cohort had been clinically misdiagnosed, and their families were subsequently able to avail of genetic counseling. In some cases, a more appropriate treatment was selected for the patient in question, or an inappropriate treatment discontinued. Our findings suggest the existence of modifier genes that may explain the incomplete penetrance of some epilepsy-related genes. We discuss possible reasons for non-diagnosis and future research directions. Further studies will be required to uncover the roles of structural variants, epimutations, and oligogenic inheritance in epilepsy, thereby providing a more complete molecular picture of this disease. In summary, given the broad phenotypic spectrum of most epilepsy-related genes, efficient genomic tools like the targeted exome sequencing panel described here are essential for early diagnosis and treatment, and should be implemented as first-tier diagnostic tools for children with epilepsy without a clear etiologic basis.
Keywords: de novo mutations; epilepsy; genetic diagnosis; incomplete penetrance; modifier genes; neurodevelopmental disorders.
Copyright © 2019 Fernández-Marmiesse, Roca, Díaz-Flores, Cantarín, Pérez-Poyato, Fontalba, Laranjeira, Quintans, Moldovan, Felgueroso, Rodríguez-Pedreira, Simón, Camacho, Quijada, Ibanez-Mico, Domingno, Benito, Calvo, Pérez-Cejas, Carrasco, Ramos, Couce, Ruiz-Falcó, Gutierrez-Solana and Martínez-Atienza.
Figures


References
-
- Babraham Bioinformatics-FastQC A Quality Control Tool for High Throughput Sequence Data (2018). Available online at: https://www.bioinformatics.babraham.ac.uk/projects/fastqc/ (accessed October 11, 2018).

