Proteomics-Based Mechanistic Investigation of Escherichia coli Inactivation by Pulsed Electric Field
- PMID: 31781086
- PMCID: PMC6857472
- DOI: 10.3389/fmicb.2019.02644
Proteomics-Based Mechanistic Investigation of Escherichia coli Inactivation by Pulsed Electric Field
Abstract
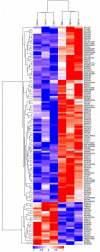
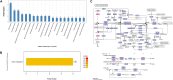
The pulsed electric field (PEF) technology has been widely applied to inactivate pathogenic bacteria in food products. Though irreversible pore formation and membrane disruption is considered to be the main contributing factor to PEF's sterilizing effects, the exact molecular mechanisms remain poorly understood. In this study, by using mass spectrometry (MS)-based label-free quantitative proteomic analysis, we compared the protein profiles of PEF-treated and untreated Escherichia coli. We identified a total of 175 differentially expressed proteins, including 52 candidates that were only detected in at least two of the three samples in one experiment group but not in the other group. Functional analysis revealed that the differential proteins were primarily involved in the regulation of cell membrane composition and integrity, stress response, as well as various metabolic processes. Quantitative reverse-transcription polymerase chain reaction (qRT-PCR) analysis was conducted on the genes of selected differential proteins at varying PEF intensities, which were known to result in different cell killing levels. The qRT-PCR data confirmed that the proteomic results could be reliably used for further data interpretation, and that the changes in the expression levels of the differential candidates were, to a large extent, caused directly by the PEF treatment. The findings of the current study offered valuable insight into PEF-induced cell inactivation.
Keywords: E. coli; cell inactivation; molecular mechanisms; proteomics; pulsed electric field.
Copyright © 2019 Liu, Zhao, Zhang, Huo, Shi, Li, Jia, Lu, Peng and Song.
Figures



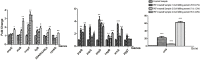
References
-
- Aronsson K., Rönner U., Borch E. (2005). Inactivation of Escherichia coli, listeria innocua and saccharomyces cerevisiae in relation to membrane permeabilization and subsequent leakage of intracellular compounds due to pulsed electric field processing. Int. J. Food Microbiol. 99 19–32. 10.1016/j.ijfoodmicro.2004.07.012 - DOI - PubMed
-
- Bolla J. R. (2014). Bacterial Multidrug Efflux Pumps: Structure, Function and Regulation. Ph.D. thesis, Iowa State University, Ames, IA.
-
- Cebrián G., Condón S., Mañas P. (2016). Influence of growth and treatment temperature on staphylococcus aureus resistance to pulsed electric fields: relationship with membrane fluidity. Innov. Food Sci. Emerg. Technol. 37 161–169. 10.1016/j.ifset.2016.08.011 - DOI
-
- Chueca B., Pagán R., García-Gonzalo D. (2015). Transcriptomic analysis of Escherichia coli MG1655 cells exposed to pulsed electric fields. Innov. Food Sci. Emerg. Technol. 29 78–86. 10.1016/j.ifset.2014.09.003 - DOI
-
- Clark J. (2006). Pulsed electric field processing. Food Technol. 60 66–67.
LinkOut - more resources
Full Text Sources

