Membrane-Specific Targeting of Tail-Anchored Proteins SECE1 and SECE2 Within Chloroplasts
- PMID: 31781139
- PMCID: PMC6857650
- DOI: 10.3389/fpls.2019.01401
Membrane-Specific Targeting of Tail-Anchored Proteins SECE1 and SECE2 Within Chloroplasts
Abstract
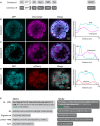
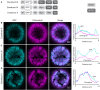
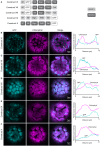
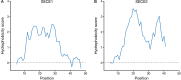
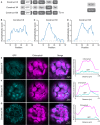
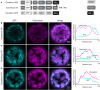
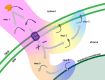
Membrane proteins that are imported into chloroplasts must be accurately targeted in order to maintain the identity and function of the highly differentiated internal membranes. Relatively little is known about the targeting information or pathways that direct proteins with transmembrane domains to either the inner envelope or thylakoids. In this study, we focused on a structurally simple class of membrane proteins, the tail-anchored proteins, which have stroma-exposed amino-terminal domains and a single transmembrane domain within 30 amino acids of the carboxy-terminus. SECE1 and SECE2 are essential tail-anchored proteins that function as components of the dual SEC translocases in chloroplasts. SECE1 localizes to the thylakoids, while SECE2 localizes to the inner envelope. We have used transient expression in Arabidopsis leaf protoplasts and confocal microscopy in combination with a domain-swapping strategy to identify regions that contain important targeting determinants. We show that membrane-specific targeting depends on features of the transmembrane domains and the short C-terminal tails. We probed the contributions of these regions to targeting processes further through site-directed mutagenesis. We show that thylakoid targeting still occurs when changes are made to the tail of SECE1, but changing residues in the tail of SECE2 abolishes inner envelope targeting. Finally, we discuss possible parallels between sorting of tail-anchored proteins in the stroma and in the cytosol.
Keywords: SEC translocase; chloroplast; inner envelope membrane; organelle biogenesis; tail-anchored protein; targeting; thylakoid.
Copyright © 2019 Anderson, Singhal and Fernandez.
Figures


Similar articles
-
The Sec2 translocase of the chloroplast inner envelope contains a unique and dedicated SECE2 component.Plant J. 2015 Nov;84(4):647-58. doi: 10.1111/tpj.13028. Epub 2015 Oct 16. Plant J. 2015. PMID: 26406904
-
Sorting of SEC translocase SCY components to different membranes in chloroplasts.J Exp Bot. 2017 Nov 2;68(18):5029-5043. doi: 10.1093/jxb/erx318. J Exp Bot. 2017. PMID: 28992187 Free PMC article.
-
Identification of Putative Substrates of SEC2, a Chloroplast Inner Envelope Translocase.Plant Physiol. 2017 Apr;173(4):2121-2137. doi: 10.1104/pp.17.00012. Epub 2017 Feb 17. Plant Physiol. 2017. PMID: 28213560 Free PMC article.
-
Two paths diverged in the stroma: targeting to dual SEC translocase systems in chloroplasts.Photosynth Res. 2018 Dec;138(3):277-287. doi: 10.1007/s11120-018-0541-9. Epub 2018 Jun 27. Photosynth Res. 2018. PMID: 29951837 Review.
-
Targeting and biogenesis of transporters and channels in chloroplast envelope membranes: Unsolved questions.Cell Calcium. 2015 Jul;58(1):122-30. doi: 10.1016/j.ceca.2014.10.012. Epub 2014 Oct 31. Cell Calcium. 2015. PMID: 25465895 Review.
Cited by
-
Structures of Get3d reveal a distinct architecture associated with the emergence of photosynthesis.J Biol Chem. 2023 Jun;299(6):104752. doi: 10.1016/j.jbc.2023.104752. Epub 2023 Apr 24. J Biol Chem. 2023. PMID: 37100288 Free PMC article.
-
Targeting signals required for protein sorting to sub-chloroplast compartments.Plant Cell Rep. 2024 Dec 26;44(1):14. doi: 10.1007/s00299-024-03409-2. Plant Cell Rep. 2024. PMID: 39724313 Review.
-
A conserved guided entry of tail-anchored pathway is involved in the trafficking of a subset of membrane proteins in Plasmodium falciparum.PLoS Pathog. 2021 Nov 15;17(11):e1009595. doi: 10.1371/journal.ppat.1009595. eCollection 2021 Nov. PLoS Pathog. 2021. PMID: 34780541 Free PMC article.
-
VaMIEL1-mediated ubiquitination of VaMYB4a orchestrates cold tolerance through integrated transcriptional and oxidative stress pathways in grapevine.Hortic Res. 2025 Mar 22;12(7):uhaf093. doi: 10.1093/hr/uhaf093. eCollection 2025 Jul. Hortic Res. 2025. PMID: 40391385 Free PMC article.
-
Protein Targeting Into the Thylakoid Membrane Through Different Pathways.Front Physiol. 2022 Jan 12;12:802057. doi: 10.3389/fphys.2021.802057. eCollection 2021. Front Physiol. 2022. PMID: 35095563 Free PMC article. Review.
References
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

