Xylem Cell Wall Formation in Pioneer Roots and Stems of Populus trichocarpa (Torr. & Gray)
- PMID: 31781142
- PMCID: PMC6861220
- DOI: 10.3389/fpls.2019.01419
Xylem Cell Wall Formation in Pioneer Roots and Stems of Populus trichocarpa (Torr. & Gray)
Abstract
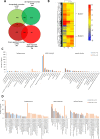
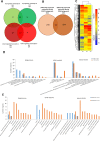
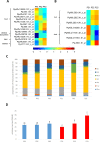
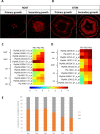
Regulation of gene expression, as determined by the genetics of the tree species, is a major factor in determining wood quality. Therefore, the identification of genes that play a role in xylogenesis is extremely important for understanding the mechanisms shaping the plant phenotype. Efforts to develop new varieties characterized by higher yield and better wood quality will greatly benefit from recognizing and understanding the complex transcriptional network underlying wood development. The present study provides a detailed comparative description of the changes that occur in genes transcription and the biosynthesis of cell-wall-related compounds during xylogenesis in Populus trichocarpa pioneer roots and stems. Even though results of microarray analysis indicated that only approximately 10% of the differentially expressed genes were common to both organs, many fundamental mechanisms were similar; e.g. the pattern of expression of genes involved in the biosynthesis of cell wall proteins, polysaccharides, and lignins. Gas chromatography time-of-flight mass spectrometry (GC-TOF-MS) shows that the composition of monosaccharides was also very similar, with an increasing amount of xylose building secondary cell wall hemicellulose and pectins, especially in the stems. While hemicellulose degradation was typical for stems, possibly due to the intensive level of cell wall lignification. Notably, the main component of lignins in roots were guiacyl units, while syringyl units were dominant in stems, where fibers are especially needed for support. Our study is the first comprehensive analysis, at the structural and molecular level, of xylogenesis in under- and aboveground tree parts, and clearly reveals the great complexity of molecular mechanisms underlying cell wall formation and modification during xylogenesis in different plant organs.
Keywords: Populus trichocarpa; cell wall biogenesis; microarrays; wood; xylogenesis.
Copyright © 2019 Marzec-Schmidt, Ludwików, Wojciechowska, Kasprowicz-Maluśki, Mucha and Bagniewska-Zadworna.
Figures
Similar articles
-
Allies or Enemies: The Role of Reactive Oxygen Species in Developmental Processes of Black Cottonwood (Populus trichocarpa).Antioxidants (Basel). 2020 Feb 27;9(3):199. doi: 10.3390/antiox9030199. Antioxidants (Basel). 2020. PMID: 32120843 Free PMC article.
-
New insights into pioneer root xylem development: evidence obtained from Populus trichocarpa plants grown under field conditions.Ann Bot. 2014 Jun;113(7):1235-47. doi: 10.1093/aob/mcu063. Epub 2014 May 8. Ann Bot. 2014. PMID: 24812251 Free PMC article.
-
Genome-wide characterization of aspartic protease (AP) gene family in Populus trichocarpa and identification of the potential PtAPs involved in wood formation.BMC Plant Biol. 2019 Jun 24;19(1):276. doi: 10.1186/s12870-019-1865-0. BMC Plant Biol. 2019. PMID: 31234799 Free PMC article.
-
Unravelling cell wall formation in the woody dicot stem.Plant Mol Biol. 2001 Sep;47(1-2):239-74. Plant Mol Biol. 2001. PMID: 11554475 Review.
-
Wood cell walls: biosynthesis, developmental dynamics and their implications for wood properties.Curr Opin Plant Biol. 2008 Jun;11(3):293-300. doi: 10.1016/j.pbi.2008.03.003. Epub 2008 Apr 21. Curr Opin Plant Biol. 2008. PMID: 18434240 Review.
Cited by
-
Promoter Cis-Element Analyses Reveal the Function of αVPE in Drought Stress Response of Arabidopsis.Biology (Basel). 2023 Mar 10;12(3):430. doi: 10.3390/biology12030430. Biology (Basel). 2023. PMID: 36979122 Free PMC article.
-
Allies or Enemies: The Role of Reactive Oxygen Species in Developmental Processes of Black Cottonwood (Populus trichocarpa).Antioxidants (Basel). 2020 Feb 27;9(3):199. doi: 10.3390/antiox9030199. Antioxidants (Basel). 2020. PMID: 32120843 Free PMC article.
-
From the Soft to the Hard: Changes in Microchemistry During Cell Wall Maturation of Walnut Shells.Front Plant Sci. 2020 Apr 21;11:466. doi: 10.3389/fpls.2020.00466. eCollection 2020. Front Plant Sci. 2020. PMID: 32431720 Free PMC article.
-
Comparative Transcriptomic Analysis of the Development of Sepal Morphology in Tomato (Solanum Lycopersicum L.).Int J Mol Sci. 2020 Aug 18;21(16):5914. doi: 10.3390/ijms21165914. Int J Mol Sci. 2020. PMID: 32824631 Free PMC article.
-
Is autophagy always a death sentence? A case study of highly selective cytoplasmic degradation during phloemogenesis.Ann Bot. 2025 Mar 13;135(4):681-696. doi: 10.1093/aob/mcae195. Ann Bot. 2025. PMID: 39497527 Free PMC article.
References
-
- Bischoff V., Nita S., Neumetzler L., Schindelasch D., Urbain A., Eshed R., et al. (2010). TRICHOME BIREFRINGENCE and Its Homolog AT5G01360 Encode Plant-Specific DUF231 Proteins Required for Cellulose Biosynthesis in Arabidopsis. Plant Physiol. 153, 590–602. 10.1104/pp.110.153320 - DOI - PMC - PubMed
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

