Integrating the Ribonucleic Acid Sequencing Data From Various Studies for Exploring the Multiple Sclerosis-Related Long Noncoding Ribonucleic Acids and Their Functions
- PMID: 31781177
- PMCID: PMC6861379
- DOI: 10.3389/fgene.2019.01136
Integrating the Ribonucleic Acid Sequencing Data From Various Studies for Exploring the Multiple Sclerosis-Related Long Noncoding Ribonucleic Acids and Their Functions
Abstract
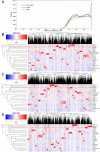
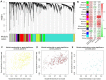
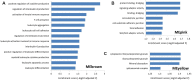
Multiple sclerosis (MS) is a chronic fatal central nervous system (CNS) disease involving in complex immunity dysfunction. Recently, long noncoding RNAs (lncRNAs) were discovered as the important regulatory factors for the pathogenesis of MS. However, these findings often cannot be repeated and confirmed by the subsequent studies. We considered that the small-scale samples or the heterogeneity among various tissues may result in the divergence of the results. Currently, RNA-seq has become a powerful approach to quantify the abundances of lncRNA transcripts. Therefore, we comprehensively collected the MS-related RNA-seq data from a variety of previous studies, and integrated these data using an expression-based meta-analysis to identify the differentially expressed lncRNA between MS patients and controls in whole samples and subgroups. Then, we performed the Jensen-Shannon (JS) divergence and cluster analysis to explore the heterogeneity and expression specificity among various tissues. Finally, we investigated the potential function of identified lncRNAs for MS using weighted gene co-expression network analysis (WGCNA) and gene set enrichment analysis (GSEA), and 5,420 MS-related lncRNAs specifically expressed in the brain tissue were identified. The subgroup analysis found a small heterogeneity of the lncRNA expression profiles between brain and blood tissues. The results of WGCNA and GSEA showed that a potential important function of lncRNAs in MS may be involved in the regulation of ribonucleoproteins and tumor necrosis factor cytokines receptors. In summary, this study provided a strategy to explore disease-related lncRNAs on genome-wide scale, and our findings will be benefit to improve the understanding of MS pathogenesis.
Keywords: function analysis; long non-coding ribonucleic acids; meta-analysis; multiple sclerosis; ribonucleic acid sequencing.
Copyright © 2019 Han, Hua, Xue and Zhu.
Figures


Similar articles
-
Genome-wide identification and analysis of the eQTL lncRNAs in multiple sclerosis based on RNA-seq data.Brief Bioinform. 2020 May 21;21(3):1023-1037. doi: 10.1093/bib/bbz036. Brief Bioinform. 2020. PMID: 31323688
-
Transcriptome analysis of lncRNA expression patterns in human congenital lung malformations.BMC Genomics. 2021 Nov 29;22(1):861. doi: 10.1186/s12864-021-08204-x. BMC Genomics. 2021. PMID: 34844556 Free PMC article.
-
Excavating novel diagnostic and prognostic long non-coding RNAs (lncRNAs) for head and neck squamous cell carcinoma: an integrated bioinformatics analysis of competing endogenous RNAs (ceRNAs) and gene co-expression networks.Bioengineered. 2021 Dec;12(2):12821-12838. doi: 10.1080/21655979.2021.2003925. Bioengineered. 2021. PMID: 34898376 Free PMC article.
-
Roles of long noncoding RNAs in brain development, functional diversification and neurodegenerative diseases.Brain Res Bull. 2013 Aug;97:69-80. doi: 10.1016/j.brainresbull.2013.06.001. Epub 2013 Jun 10. Brain Res Bull. 2013. PMID: 23756188 Review.
-
Role of long non-coding RNAs (LncRNAs) in multiple sclerosis: a brief review.Neurol Sci. 2020 Sep;41(9):2443-2451. doi: 10.1007/s10072-020-04425-2. Epub 2020 Apr 30. Neurol Sci. 2020. PMID: 32350675 Review.
Cited by
-
Meta-Analysis of RNA-Seq Datasets Identifies Novel Players in Glioblastoma.Cancers (Basel). 2022 Nov 24;14(23):5788. doi: 10.3390/cancers14235788. Cancers (Basel). 2022. PMID: 36497269 Free PMC article.
-
Identification of Novel Key Genes and Pathways in Multiple Sclerosis Based on Weighted Gene Coexpression Network Analysis and Long Noncoding RNA-Associated Competing Endogenous RNA Network.Oxid Med Cell Longev. 2022 Mar 2;2022:9328160. doi: 10.1155/2022/9328160. eCollection 2022. Oxid Med Cell Longev. 2022. PMID: 35281467 Free PMC article.
-
A Comprehensive Exploration of the Transcriptomic Landscape in Multiple Sclerosis: A Systematic Review.Int J Mol Sci. 2023 Jan 11;24(2):1448. doi: 10.3390/ijms24021448. Int J Mol Sci. 2023. PMID: 36674968 Free PMC article.
-
Comparative Transcriptional Analysis Identified Characteristic Genes and Patterns in HIV-Infected Immunological Non-Responders.Front Immunol. 2022 Jan 28;13:807890. doi: 10.3389/fimmu.2022.807890. eCollection 2022. Front Immunol. 2022. PMID: 35154126 Free PMC article.
-
Biomarkers in Rare Demyelinating Disease of the Central Nervous System.Int J Mol Sci. 2020 Nov 9;21(21):8409. doi: 10.3390/ijms21218409. Int J Mol Sci. 2020. PMID: 33182495 Free PMC article. Review.
References
-
- Angerer I. C., Hecker M., Koczan D., Roch L., Friess J., Ruge A., et al. (2018). Transcriptome profiling of peripheral blood immune cell populations in multiple sclerosis patients before and during treatment with a sphingosine-1-phosphate receptor modulator. CNS Neurosci. Ther. 24, 193–201. 10.1111/cns.12793 - DOI - PMC - PubMed
LinkOut - more resources
Full Text Sources

