Strategies for developing pregnane X receptor antagonists: Implications from metabolism to cancer
- PMID: 31782213
- PMCID: PMC7166136
- DOI: 10.1002/med.21648
Strategies for developing pregnane X receptor antagonists: Implications from metabolism to cancer
Abstract
Pregnane X receptor (PXR) is a ligand-activated nuclear receptor (NR) that was originally identified as a master regulator of xenobiotic detoxification. It regulates the expression of drug-metabolizing enzymes and transporters to control the degradation and excretion of endobiotics and xenobiotics, including therapeutic agents. The metabolism and disposition of drugs might compromise their efficacy and possibly cause drug toxicity and/or drug resistance. Because many drugs can promiscuously bind and activate PXR, PXR antagonists might have therapeutic value in preventing and overcoming drug-induced PXR-mediated drug toxicity and drug resistance. Furthermore, PXR is now known to have broader cellular functions, including the regulation of cell proliferation, and glucose and lipid metabolism. Thus, PXR might be involved in human diseases such as cancer and metabolic diseases. The importance of PXR antagonists is discussed in the context of the role of PXR in xenobiotic sensing and other disease-related pathways. This review focuses on the development of PXR antagonists, which has been hampered by the promiscuity of PXR ligand binding. However, substantial progress has been made in recent years, suggesting that it is feasible to develop selective PXR antagonists. We discuss the current status, challenges, and strategies in developing selective PXR antagonists. The strategies are based on the molecular mechanisms of antagonism in related NRs that can be applied to the design of PXR antagonists, primarily driven by structural information.
Keywords: PXR; antagonist; detoxification; small molecule; xenobiotics.
© 2019 Wiley Periodicals, Inc.
Conflict of interest statement
CONFLICT OF INTERESTS
The authors declare no conflict of interests.
Figures

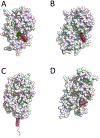
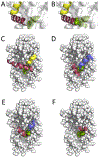
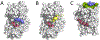

Similar articles
-
The role of pregnane X receptor (PXR) in substance metabolism.Front Endocrinol (Lausanne). 2022 Aug 16;13:959902. doi: 10.3389/fendo.2022.959902. eCollection 2022. Front Endocrinol (Lausanne). 2022. PMID: 36111293 Free PMC article. Review.
-
Mechanisms and Therapeutic Advances of PXR in Metabolic Diseases and Cancer.Int J Mol Sci. 2025 Aug 20;26(16):8029. doi: 10.3390/ijms26168029. Int J Mol Sci. 2025. PMID: 40869350 Free PMC article. Review.
-
Pregnane xenobiotic receptor in cancer pathogenesis and therapeutic response.Cancer Lett. 2013 Jan 1;328(1):1-9. doi: 10.1016/j.canlet.2012.08.030. Epub 2012 Aug 29. Cancer Lett. 2013. PMID: 22939994 Free PMC article. Review.
-
The xenobiotic receptors PXR and CAR in liver physiology, an update.Biochim Biophys Acta Mol Basis Dis. 2021 Jun 1;1867(6):166101. doi: 10.1016/j.bbadis.2021.166101. Epub 2021 Feb 15. Biochim Biophys Acta Mol Basis Dis. 2021. PMID: 33600998 Free PMC article. Review.
-
[Role of pregnane X receptor (PXR) in endobiotic metabolism].Sheng Li Xue Bao. 2019 Apr 25;71(2):311-318. Sheng Li Xue Bao. 2019. PMID: 31008491 Review. Chinese.
Cited by
-
The Nuclear Receptor PXR in Chronic Liver Disease.Cells. 2021 Dec 27;11(1):61. doi: 10.3390/cells11010061. Cells. 2021. PMID: 35011625 Free PMC article. Review.
-
Drug Mimicry: Promiscuous Receptors PXR and AhR, and Microbial Metabolite Interactions in the Intestine.Trends Pharmacol Sci. 2020 Dec;41(12):900-908. doi: 10.1016/j.tips.2020.09.013. Epub 2020 Oct 20. Trends Pharmacol Sci. 2020. PMID: 33097284 Free PMC article. Review.
-
Molecular basis of crosstalk in nuclear receptors: heterodimerization between PXR and CAR and the implication in gene regulation.Nucleic Acids Res. 2022 Apr 8;50(6):3254-3275. doi: 10.1093/nar/gkac133. Nucleic Acids Res. 2022. PMID: 35212371 Free PMC article.
-
Chemical manipulation of an activation/inhibition switch in the nuclear receptor PXR.Nat Commun. 2024 May 14;15(1):4054. doi: 10.1038/s41467-024-48472-1. Nat Commun. 2024. PMID: 38744881 Free PMC article.
-
Patchouli Alcohol Modulates the Pregnancy X Receptor/Toll-like Receptor 4/Nuclear Factor Kappa B Axis to Suppress Osteoclastogenesis.Front Pharmacol. 2021 Jun 8;12:684976. doi: 10.3389/fphar.2021.684976. eCollection 2021. Front Pharmacol. 2021. PMID: 34177594 Free PMC article.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical

