The influence of DNA methylation on monoallelic expression
- PMID: 31782494
- PMCID: PMC6923323
- DOI: 10.1042/EBC20190034
The influence of DNA methylation on monoallelic expression
Abstract
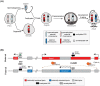
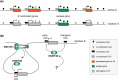
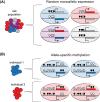
Monoallelic gene expression occurs in diploid cells when only one of the two alleles of a gene is active. There are three main classes of genes that display monoallelic expression in mammalian genomes: (1) imprinted genes that are monoallelically expressed in a parent-of-origin dependent manner; (2) X-linked genes that undergo random X-chromosome inactivation in female cells; (3) random monoallelically expressed single and clustered genes located on autosomes. The heritability of monoallelic expression patterns during cell divisions implies that epigenetic mechanisms are involved in the cellular memory of these expression states. Among these, methylation of CpG sites on DNA is one of the best described modification to explain somatic inheritance. Here, we discuss the relevance of DNA methylation for the establishment and maintenance of monoallelic expression patterns among these three groups of genes, and how this is intrinsically linked to development and cellular states.
Keywords: DNA methylation; X-chromosome inactivation; epigenetics; imprinting; monoallelic expression.
© 2019 The Author(s).
Conflict of interest statement
The authors declare that there are no competing interests associated with the manuscript.
Figures
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

