Circular RNA hsa_circ_001895 serves as a sponge of microRNA-296-5p to promote clear cell renal cell carcinoma progression by regulating SOX12
- PMID: 31782868
- PMCID: PMC7004537
- DOI: 10.1111/cas.14261
Circular RNA hsa_circ_001895 serves as a sponge of microRNA-296-5p to promote clear cell renal cell carcinoma progression by regulating SOX12
Erratum in
-
Correction to Circular RNA hsa_circ_001895 serves as a sponge of microRNA-296-5p to promote clear cell renal cell carcinoma progression by regulating SOX12.Cancer Sci. 2023 Nov;114(11):4475. doi: 10.1111/cas.15968. Epub 2023 Oct 2. Cancer Sci. 2023. PMID: 37782017 Free PMC article. No abstract available.
Abstract
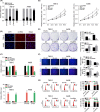
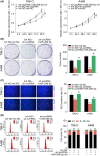
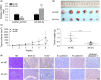
There is an urgent need to find novel potential therapeutic targets for the diagnosis and treatment of clear cell renal cell carcinoma (ccRCC) due to its highly invasive ability as a common urological malignant tumor. Circular RNAs (circRNAs) have been indicated as potentially critical mediators in various types of tumor progression. We first used qRT-PCR analysis to find dysregulated circRNAs in ccRCC. A novel circRNA, hsa_circ_001895, was upregulated in ccRCC specimens and associated with metastatic properties of ccRCC. However, the tumorigenic mechanism of hsa_circ_001895 on ccRCC is yet to be found. We first indicated that hsa_circ_001895 predicted a poor prognosis in ccRCC patients. Additionally, overexpression of hsa_circ_001895 not only promoted cell proliferation, invasion and migration of ccRCC, but also inhibited cell apoptosis, whereas hsa_circ_001895 knockdown reversed the effect on ccRCC progression. In vivo s.c. xenotransplanted tumor model also showed that silencing hsa_circ_001895 could suppress in vivo ccRCC growth. Mechanistically, hsa_circ_001895 directly binds with microRNA (miR)-296-5p and inhibits its expression. Moreover, sex determining region Y (SRY)-box 12 (SOX12) was identified as a target of miR-296-5p, the expression of which was suppressed by miR-296-5p. Notably, the inhibitory effect of hsa_circ_001895 on ccRCC progression was reversed by miR-296-5p inhibitor. In general, our findings indicated that hsa_circ_001895 may sponge miR-296-5p and promote SOX12 expression, which is the underlying mechanism of hsa_circ_001895-induced ccRCC progression.
Keywords: SOX12; clear cell renal cell carcinoma; hsa_circ_001895; miR-296-5p; progression.
© 2019 The Authors. Cancer Science published by John Wiley & Sons Australia, Ltd on behalf of Japanese Cancer Association.
Conflict of interest statement
Authors declare no conflicts of interest for this article.
Figures





Similar articles
-
circCHST15 is a novel prognostic biomarker that promotes clear cell renal cell carcinoma cell proliferation and metastasis through the miR-125a-5p/EIF4EBP1 axis.Mol Cancer. 2021 Dec 18;20(1):169. doi: 10.1186/s12943-021-01449-w. Mol Cancer. 2021. PMID: 34922539 Free PMC article.
-
Hsa_circ_0085576 promotes clear cell renal cell carcinoma tumorigenesis and metastasis through the miR-498/YAP1 axis.Aging (Albany NY). 2020 Jun 15;12(12):11530-11549. doi: 10.18632/aging.103300. Epub 2020 Jun 15. Aging (Albany NY). 2020. PMID: 32541093 Free PMC article.
-
Circ_0003520/miR-205-5p/CUL4B Axis Drives the Progression of Clear Cell Renal Carcinoma.J Biochem Mol Toxicol. 2025 May;39(5):e70263. doi: 10.1002/jbt.70263. J Biochem Mol Toxicol. 2025. PMID: 40320861
-
Emerging Role of Circular RNAs in the Pathogenesis of Retinoblastoma.Ophthalmic Res. 2024;67(1):51-61. doi: 10.1159/000535329. Epub 2023 Dec 18. Ophthalmic Res. 2024. PMID: 38109867 Review.
-
Non-coding transcriptome profiles in clear-cell renal cell carcinoma.Nat Rev Urol. 2025 Mar;22(3):151-174. doi: 10.1038/s41585-024-00926-3. Epub 2024 Sep 6. Nat Rev Urol. 2025. PMID: 39242964 Review.
Cited by
-
New insight into circRNAs: characterization, strategies, and biomedical applications.Exp Hematol Oncol. 2023 Oct 12;12(1):91. doi: 10.1186/s40164-023-00451-w. Exp Hematol Oncol. 2023. PMID: 37828589 Free PMC article. Review.
-
Circ-APBB1IP as a Prognostic Biomarker Promotes Clear Cell Renal Cell Carcinoma Progression Through The ERK1/2 Signaling Pathway.Int J Med Sci. 2020 May 18;17(9):1177-1186. doi: 10.7150/ijms.44550. eCollection 2020. Int J Med Sci. 2020. PMID: 32547313 Free PMC article.
-
Hsa_circRNA_000166 Promotes Cell Proliferation, Migration and Invasion by Regulating miR-330-5p/ELK1 in Colon Cancer.Onco Targets Ther. 2020 Jun 12;13:5529-5539. doi: 10.2147/OTT.S243795. eCollection 2020. Onco Targets Ther. 2020. Retraction in: Onco Targets Ther. 2024 Apr 03;17:285-286. doi: 10.2147/OTT.S471178. PMID: 32606768 Free PMC article. Retracted.
-
CircRNAs as Novel Biomarkers and Therapeutic Targets in Renal Cell Carcinoma.Front Mol Biosci. 2022 Feb 11;9:833079. doi: 10.3389/fmolb.2022.833079. eCollection 2022. Front Mol Biosci. 2022. PMID: 35223991 Free PMC article. Review.
-
Circular RNAs as prognostic and diagnostic biomarkers in renal cell carcinoma.J Clin Lab Anal. 2022 Oct;36(10):e24670. doi: 10.1002/jcla.24670. Epub 2022 Aug 21. J Clin Lab Anal. 2022. PMID: 35989533 Free PMC article. Review.
References
-
- Ghali MGZ. Role of the medullary lateral tegmental field in sympathetic control. J Integr Neurosci. 2017;16:189‐208. - PubMed
-
- Jemal A, Bray F, Center MM, Ferlay J, Ward E, Forman D. Global cancer statistics. CA Cancer J Clin. 2011;61(2):69‐90. - PubMed
-
- Siegel RL, Miller KD, Jemal A. Cancer statistics, 2017. CA Cancer J Clin. 2017;67(1):7‐30. - PubMed
-
- Powles T, Staehler M, Ljungberg B, et al. European Association of Urology Guidelines for clear cell renal cancers that are resistant to vascular endothelial growth factor receptor‐targeted therapy. Eur Urol. 2016;70(5):705‐706. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical

