Crystal Structure of a GH3 β-Glucosidase from the Thermophilic Fungus Chaetomium thermophilum
- PMID: 31783503
- PMCID: PMC6929035
- DOI: 10.3390/ijms20235962
Crystal Structure of a GH3 β-Glucosidase from the Thermophilic Fungus Chaetomium thermophilum
Abstract
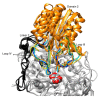
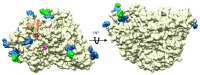
Beta-glucosidases (β-glucosidases) have attracted considerable attention in recent years for use in various biotechnological applications. They are also essential enzymes for lignocellulose degradation in biofuel production. However, cost-effective biomass conversion requires the use of highly efficient enzymes. Thus, the search for new enzymes as better alternatives of the currently available enzyme preparations is highly important. Thermophilic fungi are nowadays considered as a promising source of enzymes with improved stability. Here, the crystal structure of a family GH3 β-glucosidase from the thermophilic fungus Chaetomium thermophilum (CtBGL) was determined at a resolution of 2.99 Å. The structure showed the three-domain architecture found in other β-glucosidases with variations in loops and linker regions. The active site catalytic residues in CtBGL were identified as Asp287 (nucleophile) and Glu517 (acid/base). Structural comparison of CtBGL with Protein Data Bank (PDB)-deposited structures revealed variations among glycosylated Asn residues. The enzyme displayed moderate glycosylation compared to other GH3 family β-glucosidases with similar structure. A new glycosylation site at position Asn504 was identified in CtBGL. Moreover, comparison with respect to several thermostability parameters suggested that glycosylation and charged residues involved in electrostatic interactions may contribute to the stability of the enzyme at elevated temperatures. The reported CtBGL structure provides additional insights into the family GH3 enzymes and could offer new ideas for further improvements in β-glucosidases for more efficient use in biotechnological applications regarding cellulose degradation.
Keywords: Chaetomium thermophilum; cellulose degradation; fungal enzymes; glycoside hydrolase; protein structure; thermophilic fungus; β-glucosidases.
Conflict of interest statement
The authors declare no conflicts of interest.
Figures



Similar articles
-
Expression of Two Novel β-Glucosidases from Chaetomium atrobrunneum in Trichoderma reesei and Characterization of the Heterologous Protein Products.Mol Biotechnol. 2016 Dec;58(12):821-831. doi: 10.1007/s12033-016-9981-7. Mol Biotechnol. 2016. PMID: 27714589
-
Structural and functional studies of the glycoside hydrolase family 3 β-glucosidase Cel3A from the moderately thermophilic fungus Rasamsonia emersonii.Acta Crystallogr D Struct Biol. 2016 Jul;72(Pt 7):860-70. doi: 10.1107/S2059798316008482. Epub 2016 Jun 23. Acta Crystallogr D Struct Biol. 2016. PMID: 27377383 Free PMC article.
-
Three-dimensional structures of two heavily N-glycosylated Aspergillus sp. family GH3 β-D-glucosidases.Acta Crystallogr D Struct Biol. 2016 Feb;72(Pt 2):254-65. doi: 10.1107/S2059798315024237. Epub 2016 Jan 28. Acta Crystallogr D Struct Biol. 2016. PMID: 26894673 Free PMC article.
-
Unconventional β-Glucosidases: A Promising Biocatalyst for Industrial Biotechnology.Appl Biochem Biotechnol. 2021 Sep;193(9):2993-3016. doi: 10.1007/s12010-021-03568-y. Epub 2021 Apr 19. Appl Biochem Biotechnol. 2021. PMID: 33871765 Review.
-
A glucotolerant β-glucosidase from the fungus Talaromyces amestolkiae and its conversion into a glycosynthase for glycosylation of phenolic compounds.Microb Cell Fact. 2020 Jun 10;19(1):127. doi: 10.1186/s12934-020-01386-1. Microb Cell Fact. 2020. PMID: 32522206 Free PMC article. Review.
Cited by
-
Application of Immobilized β-Glucosidase from Candida boidinii in the Hydrolysis of Delignified Sugarcane Bagasse.Indian J Microbiol. 2024 Jun;64(2):650-670. doi: 10.1007/s12088-024-01223-8. Epub 2024 Mar 5. Indian J Microbiol. 2024. PMID: 39010988 Free PMC article.
-
Structural basis of EHEP-mediated offense against phlorotannin-induced defense from brown algae to protect akuBGL activity.Elife. 2023 Nov 1;12:RP88939. doi: 10.7554/eLife.88939. Elife. 2023. PMID: 37910430 Free PMC article.
-
Profiling of the β-glucosidases identified in the genome of Penicillium funiculosum: insights from genomics, transcriptomics, proteomics, and homology-modeling studies.Appl Environ Microbiol. 2023 Sep 28;89(9):e0070423. doi: 10.1128/aem.00704-23. Epub 2023 Aug 23. Appl Environ Microbiol. 2023. PMID: 37610233 Free PMC article.
-
Carbohydrate-Active Enzymes: Structure, Activity, and Reaction Products.Int J Mol Sci. 2020 Apr 15;21(8):2727. doi: 10.3390/ijms21082727. Int J Mol Sci. 2020. PMID: 32326403 Free PMC article.
-
The evolutionary advantage of an aromatic clamp in plant family 3 glycoside exo-hydrolases.Nat Commun. 2022 Sep 23;13(1):5577. doi: 10.1038/s41467-022-33180-5. Nat Commun. 2022. PMID: 36151080 Free PMC article.
References
-
- Gudmundsson M., Hansson H., Karkehabadi S., Larsson A., Stals I., Kim S., Sunux S., Fujdala M., Larenas E., Kaper T., et al. Structural and functional studies of the glycoside hydrolase family 3 β-glucosidase Cel3A from the moderately thermophilic fungus Rasamsonia emersonii. Acta Crystallogr. Sect. D Struct. Biol. 2016;72:860–870. doi: 10.1107/S2059798316008482. - DOI - PMC - PubMed
-
- Karkehabadi S., Hansson H., Mikkelsen N.E., Kim S., Kaper T., Sandgren M., Gudmundsson M. Structural studies of a glycoside hydrolase family 3 β-glucosidase from the model fungus Neurospora crassa. Acta Crystallogr. Sect. F Struct. Biol. Commun. 2018;74:787–796. doi: 10.1107/S2053230X18015662. - DOI - PMC - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

