Comparative genomics of Mycoplasma pneumoniae isolated from children with pneumonia: South Korea, 2010-2016
- PMID: 31783732
- PMCID: PMC6884898
- DOI: 10.1186/s12864-019-6306-9
Comparative genomics of Mycoplasma pneumoniae isolated from children with pneumonia: South Korea, 2010-2016
Abstract
Background: Mycoplasma pneumoniae is a common cause of respiratory tract infections in children and adults. This study applied high-throughput whole genome sequencing (WGS) technologies to analyze the genomes of 30 M. pneumoniae strains isolated from children with pneumonia in South Korea during the two epidemics from 2010 to 2016 in comparison with a global collection of 48 M. pneumoniae strains which includes seven countries ranging from 1944 to 2017.
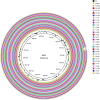
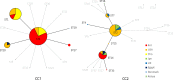
Results: The 30 Korean strains had approximately 40% GC content and ranged from 815,686 to 818,669 base pairs, coding for a total of 809 to 828 genes. Overall, BRIG revealed 99% to > 99% similarity among strains. The genomic similarity dropped to approximately 95% in the P1 type 2 strains when aligned to the reference M129 genome, which corresponded to the region of the p1 gene. MAUVE detected four subtype-specific insertions (three in P1 type 1 and one in P1 type 2), of which were all hypothetical proteins except one tRNA insertion in all P1 type 1 strains. The phylogenetic associations of 30 strains were generally consistent with the multilocus sequence typing results. The phylogenetic tree constructed with 78 genomes including 30 genomes from Korea formed two clusters and further divided into two sub-clusters. eBURST analysis revealed two clonal complexes according to P1 typing results showing higher diversity among P1 type 2 strains.
Conclusions: The comparative whole genome approach was able to define high genetic identity, unique structural diversity, and phylogenetic associations among the 78 M. pneumoniae strains isolated worldwide.
Keywords: Comparative genomics; Mycoplasma pneumoniae; Whole genome analysis.
Conflict of interest statement
The authors declare that they have no competing interests.
Figures


Similar articles
-
Comparative genomic sequencing to characterize Mycoplasma pneumoniae genome, typing, and drug resistance.Microbiol Spectr. 2024 Aug 6;12(8):e0361523. doi: 10.1128/spectrum.03615-23. Epub 2024 Jun 21. Microbiol Spectr. 2024. PMID: 38904371 Free PMC article.
-
Molecular characterization of genomic DNA in mycoplasma pneumoniae strains isolated from serious mycoplasma pneumonia cases in 2016, Yunnan, China.Infect Genet Evol. 2018 Mar;58:125-134. doi: 10.1016/j.meegid.2017.12.020. Epub 2017 Dec 22. Infect Genet Evol. 2018. PMID: 29275190
-
Molecular Typing of Mycoplasma pneumoniae Strains in Sweden from 1996 to 2017 and the Emergence of a New P1 Cytadhesin Gene, Variant 2e.J Clin Microbiol. 2019 May 24;57(6):e00049-19. doi: 10.1128/JCM.00049-19. Print 2019 Jun. J Clin Microbiol. 2019. PMID: 30918047 Free PMC article.
-
[The genetic view of Mycoplasma infections].Nihon Rinsho. 2003 Mar;61 Suppl 3:772-8. Nihon Rinsho. 2003. PMID: 12718063 Review. Japanese. No abstract available.
-
Overview of antimicrobial options for Mycoplasma pneumoniae pneumonia: focus on macrolide resistance.Clin Respir J. 2017 Jul;11(4):419-429. doi: 10.1111/crj.12379. Epub 2015 Oct 13. Clin Respir J. 2017. PMID: 26365811 Review.
Cited by
-
Detection of human IgG antibodies against Mycoplasma genitalium using a recombinant MG075 antigen.J Clin Microbiol. 2025 May 14;63(5):e0187624. doi: 10.1128/jcm.01876-24. Epub 2025 Apr 23. J Clin Microbiol. 2025. PMID: 40265939 Free PMC article.
-
Molecular Tools for Typing Mycoplasma pneumoniae and Mycoplasma genitalium.Front Microbiol. 2022 Jun 2;13:904494. doi: 10.3389/fmicb.2022.904494. eCollection 2022. Front Microbiol. 2022. PMID: 35722324 Free PMC article. Review.
-
Genotyping of Mycoplasma pneumoniae strains isolated in Japan during 2019 and 2020: spread of p1 gene type 2c and 2j variant strains.Front Microbiol. 2023 Jun 19;14:1202357. doi: 10.3389/fmicb.2023.1202357. eCollection 2023. Front Microbiol. 2023. PMID: 37405159 Free PMC article.
-
Genetic factors driving the Mycoplasma pneumoniae outbreak among children post-COVID-19 in China: a whole genome analysis.Lancet Reg Health West Pac. 2025 May 13;59:101578. doi: 10.1016/j.lanwpc.2025.101578. eCollection 2025 Jun. Lancet Reg Health West Pac. 2025. PMID: 40487843 Free PMC article.
-
Advances in adhesion-related pathogenesis in Mycoplasma pneumoniae infection.Front Microbiol. 2025 Jul 23;16:1613760. doi: 10.3389/fmicb.2025.1613760. eCollection 2025. Front Microbiol. 2025. PMID: 40771685 Free PMC article. Review.
References
-
- Spuesens EB, Fraaij PL, Visser EG, Hoogenboezem T, Hop WC, van Adrichem LN, et al. Carriage of Mycoplasma pneumoniae in the upper respiratory tract of symptomatic and asymptomatic children: an observational study. PLoS Med. 2013;10(5):e1001444. doi: 10.1371/journal.pmed.1001444. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

