SIPA1L2 controls trafficking and local signaling of TrkB-containing amphisomes at presynaptic terminals
- PMID: 31784514
- PMCID: PMC6884526
- DOI: 10.1038/s41467-019-13224-z
SIPA1L2 controls trafficking and local signaling of TrkB-containing amphisomes at presynaptic terminals
Abstract
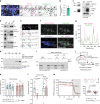
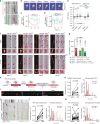
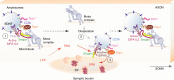
Amphisomes are organelles of the autophagy pathway that result from the fusion of autophagosomes with late endosomes. While biogenesis of autophagosomes and late endosomes occurs continuously at axon terminals, non-degradative roles of autophagy at boutons are barely described. Here, we show that in neurons BDNF/TrkB traffick in amphisomes that signal locally at presynaptic boutons during retrograde transport to the soma. This is orchestrated by the Rap GTPase-activating (RapGAP) protein SIPA1L2, which connects TrkB amphisomes to a dynein motor. The autophagosomal protein LC3 regulates RapGAP activity of SIPA1L2 and controls retrograde trafficking and local signaling of TrkB. Following induction of presynaptic plasticity, amphisomes dissociate from dynein at boutons enabling local signaling and promoting transmitter release. Accordingly, sipa1l2 knockout mice show impaired BDNF-dependent presynaptic plasticity. Taken together, the data suggest that in hippocampal neurons, TrkB-signaling endosomes are in fact amphisomes that during retrograde transport have local signaling capacity in the context of presynaptic plasticity.
Conflict of interest statement
The authors declare no competing interests.
Figures






References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
Miscellaneous

