Myeloid lineage enhancers drive oncogene synergy in CEBPA/CSF3R mutant acute myeloid leukemia
- PMID: 31784538
- PMCID: PMC6884457
- DOI: 10.1038/s41467-019-13364-2
Myeloid lineage enhancers drive oncogene synergy in CEBPA/CSF3R mutant acute myeloid leukemia
Erratum in
-
Author Correction: Myeloid lineage enhancers drive oncogene synergy in CEBPA/CSF3R mutant acute myeloid leukemia.Nat Commun. 2022 Jun 16;13(1):3471. doi: 10.1038/s41467-022-30987-0. Nat Commun. 2022. PMID: 35710794 Free PMC article. No abstract available.
Abstract
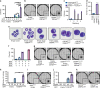
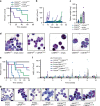
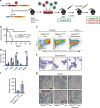
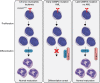
Acute Myeloid Leukemia (AML) develops due to the acquisition of mutations from multiple functional classes. Here, we demonstrate that activating mutations in the granulocyte colony stimulating factor receptor (CSF3R), cooperate with loss of function mutations in the transcription factor CEBPA to promote acute leukemia development. The interaction between these distinct classes of mutations occurs at the level of myeloid lineage enhancers where mutant CEBPA prevents activation of a subset of differentiation associated enhancers. To confirm this enhancer-dependent mechanism, we demonstrate that CEBPA mutations must occur as the initial event in AML initiation. This improved mechanistic understanding will facilitate therapeutic development targeting the intersection of oncogene cooperativity.
Conflict of interest statement
B.J.D. potential competing interests—Consultant: Monojul, Patient True Talk; SAB: Aileron Therapeutics, ALLCRON, Cepheid, Gilead Sciences, Vivid Biosciences, Celgene & Baxalta (inactive); SAB & Stock: Aptose Biosciences, Blueprint Medicines, Beta Cat, GRAIL, Third Coast Therapeutics, CTI BioPharma (inactive); Scientific Founder & Stock: MolecularMD; Board of Directors & Stock: Amgen; Board of Directors: Burroughs Wellcome Fund, CureOne; Joint Steering Committee: Beat AML LLS; Clinical Trial Funding: Novartis, Bristol-Myers Squibb, Pfizer; Royalties: OHSU #606-Novartis exclusive license, OHSU #2573; Dana-Farber Cancer Institute #2063-Merck exclusive license. The remaining authors declare no competing interests.
Figures




References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Research Materials

