The mycolic acid reductase Rv2509 has distinct structural motifs and is essential for growth in slow-growing mycobacteria
- PMID: 31785114
- PMCID: PMC7065075
- DOI: 10.1111/mmi.14437
The mycolic acid reductase Rv2509 has distinct structural motifs and is essential for growth in slow-growing mycobacteria
Abstract
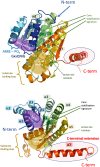
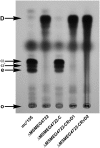
The final step in mycolic acid biosynthesis in Mycobacterium tuberculosis is catalysed by mycolyl reductase encoded by the Rv2509 gene. Sequence analysis and homology modelling indicate that Rv2509 belongs to the short-chain fatty acid dehydrogenase/reductase (SDR) family, but with some distinct features that warrant its classification as belonging to a novel family of short-chain dehydrogenases. In particular, the predicted structure revealed a unique α-helical C-terminal region which we demonstrated to be essential for Rv2509 function, though this region did not seem to play any role in protein stabilisation or oligomerisation. We also show that unlike the M. smegmatis homologue which was not essential for growth, Rv2509 was an essential gene in slow-growing mycobacteria. A knockdown strain of the BCG2529 gene, the Rv2509 homologue in Mycobacterium bovis BCG, was unable to grow following the conditional depletion of BCG2529. This conditional depletion also led to a reduction of mature mycolic acid production and accumulation of intermediates derived from 3-oxo-mycolate precursors. Our studies demonstrate novel features of the mycolyl reductase Rv2509 and outline its role in mycobacterial growth, highlighting its potential as a new target for therapies.
Keywords: Mycobacterium; dehydrogenase; mycolic acid; reductase; tuberculosis.
© 2019 The Authors. Molecular Microbiology published by John Wiley & Sons Ltd.
Figures


Similar articles
-
The reductase that catalyzes mycolic motif synthesis is required for efficient attachment of mycolic acids to arabinogalactan.J Biol Chem. 2007 Apr 13;282(15):11000-8. doi: 10.1074/jbc.M608686200. Epub 2007 Feb 17. J Biol Chem. 2007. PMID: 17308303
-
Loss of a mycobacterial gene encoding a reductase leads to an altered cell wall containing beta-oxo-mycolic acid analogs and accumulation of ketones.Chem Biol. 2008 Sep 22;15(9):930-9. doi: 10.1016/j.chembiol.2008.07.007. Chem Biol. 2008. PMID: 18804030 Free PMC article.
-
Phosphorylation of the Mycobacterium tuberculosis beta-ketoacyl-acyl carrier protein reductase MabA regulates mycolic acid biosynthesis.J Biol Chem. 2010 Apr 23;285(17):12714-25. doi: 10.1074/jbc.M110.105189. Epub 2010 Feb 23. J Biol Chem. 2010. PMID: 20178986 Free PMC article.
-
Pathway to synthesis and processing of mycolic acids in Mycobacterium tuberculosis.Clin Microbiol Rev. 2005 Jan;18(1):81-101. doi: 10.1128/CMR.18.1.81-101.2005. Clin Microbiol Rev. 2005. PMID: 15653820 Free PMC article. Review.
-
The Molecular Genetics of Mycolic Acid Biosynthesis.Microbiol Spectr. 2014 Aug;2(4):MGM2-0003-2013. doi: 10.1128/microbiolspec.MGM2-0003-2013. Microbiol Spectr. 2014. PMID: 26104214 Review.
Cited by
-
Phylogenomic Reappraisal of Fatty Acid Biosynthesis, Mycolic Acid Biosynthesis and Clinical Relevance Among Members of the Genus Corynebacterium.Front Microbiol. 2021 Dec 23;12:802532. doi: 10.3389/fmicb.2021.802532. eCollection 2021. Front Microbiol. 2021. PMID: 35003033 Free PMC article.
-
Rv0687 a Putative Short-Chain Dehydrogenase Is Required for In Vitro and In Vivo Survival of Mycobacterium tuberculosis.Int J Mol Sci. 2024 Jul 18;25(14):7862. doi: 10.3390/ijms25147862. Int J Mol Sci. 2024. PMID: 39063103 Free PMC article.
References
-
- Bardarov, S. , Bardarov, S., Jr. , Pavelka, M. S., Jr. , Sambandamurthy, V. , Larsen, M. , Tufariello, J. , … Jacobs, W. R., Jr. (2002). Specialized transduction: An efficient method for generating marked and unmarked targeted gene disruptions in Mycobacterium tuberculosis, M. bovis BCG and M. smegmatis . Microbiology, 148, 3007–3017. 10.1099/00221287-148-10-3007 - DOI - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

