DeepEP: a deep learning framework for identifying essential proteins
- PMID: 31787076
- PMCID: PMC6886168
- DOI: 10.1186/s12859-019-3076-y
DeepEP: a deep learning framework for identifying essential proteins
Abstract
Background: Essential proteins are crucial for cellular life and thus, identification of essential proteins is an important topic and a challenging problem for researchers. Recently lots of computational approaches have been proposed to handle this problem. However, traditional centrality methods cannot fully represent the topological features of biological networks. In addition, identifying essential proteins is an imbalanced learning problem; but few current shallow machine learning-based methods are designed to handle the imbalanced characteristics.
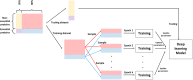
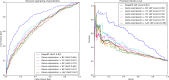
Results: We develop DeepEP based on a deep learning framework that uses the node2vec technique, multi-scale convolutional neural networks and a sampling technique to identify essential proteins. In DeepEP, the node2vec technique is applied to automatically learn topological and semantic features for each protein in protein-protein interaction (PPI) network. Gene expression profiles are treated as images and multi-scale convolutional neural networks are applied to extract their patterns. In addition, DeepEP uses a sampling method to alleviate the imbalanced characteristics. The sampling method samples the same number of the majority and minority samples in a training epoch, which is not biased to any class in training process. The experimental results show that DeepEP outperforms traditional centrality methods. Moreover, DeepEP is better than shallow machine learning-based methods. Detailed analyses show that the dense vectors which are generated by node2vec technique contribute a lot to the improved performance. It is clear that the node2vec technique effectively captures the topological and semantic properties of PPI network. The sampling method also improves the performance of identifying essential proteins.
Conclusion: We demonstrate that DeepEP improves the prediction performance by integrating multiple deep learning techniques and a sampling method. DeepEP is more effective than existing methods.
Keywords: Deep learning; Identifying essential proteins; Imbalanced learning; Multi-scale convolutional neural networks; Protein-protein interaction network; node2vec.
Conflict of interest statement
The authors declare that they have no competing interests.
Figures



Similar articles
-
DeepHE: Accurately predicting human essential genes based on deep learning.PLoS Comput Biol. 2020 Sep 16;16(9):e1008229. doi: 10.1371/journal.pcbi.1008229. eCollection 2020 Sep. PLoS Comput Biol. 2020. PMID: 32936825 Free PMC article.
-
A deep learning framework for identifying essential proteins based on multiple biological information.BMC Bioinformatics. 2022 Aug 4;23(1):318. doi: 10.1186/s12859-022-04868-8. BMC Bioinformatics. 2022. PMID: 35927611 Free PMC article.
-
A Deep Learning Framework for Identifying Essential Proteins by Integrating Multiple Types of Biological Information.IEEE/ACM Trans Comput Biol Bioinform. 2021 Jan-Feb;18(1):296-305. doi: 10.1109/TCBB.2019.2897679. Epub 2021 Feb 3. IEEE/ACM Trans Comput Biol Bioinform. 2021. PMID: 30736002
-
Convolutional Neural Networks for ATC Classification.Curr Pharm Des. 2018;24(34):4007-4012. doi: 10.2174/1381612824666181112113438. Curr Pharm Des. 2018. PMID: 30417778 Review.
-
Application of deep learning methods in biological networks.Brief Bioinform. 2021 Mar 22;22(2):1902-1917. doi: 10.1093/bib/bbaa043. Brief Bioinform. 2021. PMID: 32363401 Review.
Cited by
-
Biological network analysis with deep learning.Brief Bioinform. 2021 Mar 22;22(2):1515-1530. doi: 10.1093/bib/bbaa257. Brief Bioinform. 2021. PMID: 33169146 Free PMC article. Review.
-
'Bingo'-a large language model- and graph neural network-based workflow for the prediction of essential genes from protein data.Brief Bioinform. 2023 Nov 22;25(1):bbad472. doi: 10.1093/bib/bbad472. Brief Bioinform. 2023. PMID: 38152979 Free PMC article.
-
Untangling the Context-Specificity of Essential Genes by Means of Machine Learning: A Constructive Experience.Biomolecules. 2023 Dec 22;14(1):18. doi: 10.3390/biom14010018. Biomolecules. 2023. PMID: 38254618 Free PMC article.
-
DeepHE: Accurately predicting human essential genes based on deep learning.PLoS Comput Biol. 2020 Sep 16;16(9):e1008229. doi: 10.1371/journal.pcbi.1008229. eCollection 2020 Sep. PLoS Comput Biol. 2020. PMID: 32936825 Free PMC article.
-
Elucidating the functional roles of prokaryotic proteins using big data and artificial intelligence.FEMS Microbiol Rev. 2023 Jan 16;47(1):fuad003. doi: 10.1093/femsre/fuad003. FEMS Microbiol Rev. 2023. PMID: 36725215 Free PMC article. Review.
References
-
- Roemer T, Jiang B, Davison J, Ketela T, Veillette K, Breton A, Tandia F, Linteau A, Sillaots S, Marta C. Large-scale essential gene identification in Candida albicans and applications to antifungal drug discovery. Mol Microbiol. 2003;50(1):167–181. doi: 10.1046/j.1365-2958.2003.03697.x. - DOI - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

